DNA ultra-sensitive quantification, a technology for studying HIV unintegrated linear DNA
- PMID: 37159665
- PMCID: PMC10162948
- DOI: 10.1016/j.crmeth.2023.100443
DNA ultra-sensitive quantification, a technology for studying HIV unintegrated linear DNA
Abstract
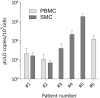
Unintegrated HIV DNA represents between 20% and 35% of the total viral DNA in infected patients. Only the linear forms (unintegrated linear DNAs [ULDs]) can be substrates for integration and for the completion of a full viral cycle. In quiescent cells, these ULDs may be responsible for pre-integrative latency. However, their detection remains difficult due to the lack of specificity and sensitivity of existing techniques. We developed an ultra-sensitive, specific, and high-throughput technology for ULD quantification called DUSQ (DNA ultra-sensitive quantification) combining linker-mediated PCR and next-generation sequencing (NGS) using molecular barcodes. Studying cells with different activity levels, we determined that the ULD half-life goes up to 11 days in resting CD4+ T cells. Finally, we were able to quantify ULDs in samples from patients infected with HIV-1, providing a proof of concept for the use of DUSQ in vivo to track pre-integrative latency. DUSQ can be adapted to the detection of other rare DNA molecules.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

