A ribosomal gene panel predicting a novel synthetic lethality in non-BRCAness tumors
- PMID: 37160887
- PMCID: PMC10170152
- DOI: 10.1038/s41392-023-01401-y
A ribosomal gene panel predicting a novel synthetic lethality in non-BRCAness tumors
Abstract
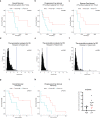
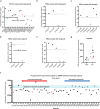
Poly (ADP-ribose) polymerase (PARP) inhibitors are one of the most exciting classes of targeted therapy agents for cancers with homologous recombination (HR) deficiency. However, many patients without apparent HR defects also respond well to PARP inhibitors/cisplatin. The biomarker responsible for this mechanism remains unclear. Here, we identified a set of ribosomal genes that predict response to PARP inhibitors/cisplatin in HR-proficient patients. PARP inhibitor/cisplatin selectively eliminates cells with high expression of the eight genes in the identified panel via DNA damage (ATM) signaling-induced pro-apoptotic ribosomal stress, which along with ATM signaling-induced pro-survival HR repair constitutes a new model to balance the cell fate in response to DNA damage. Therefore, the combined examination of the gene panel along with HR status would allow for more precise predictions of clinical response to PARP inhibitor/cisplatin. The gene panel as an independent biomarker was validated by multiple published clinical datasets, as well as by an ovarian cancer organoids library we established. More importantly, its predictive value was further verified in a cohort of PARP inhibitor-treated ovarian cancer patients with both RNA-seq and WGS data. Furthermore, we identified several marketed drugs capable of upregulating the expression of the genes in the panel without causing HR deficiency in PARP inhibitor/cisplatin-resistant cell lines. These drugs enhance PARP inhibitor/cisplatin sensitivity in both intrinsically resistant organoids and cell lines with acquired resistance. Together, our study identifies a marker gene panel for HR-proficient patients and reveals a broader application of PARP inhibitor/cisplatin in cancer therapy.
© 2023. The Author(s).
Conflict of interest statement
A patent is pending for J-S.Z., C.Z., and Q.G. Patent applicant: J-S.Z.; the name of inventors: Zhang Jin-San, Zhang Chao, Guo Qiang; application number: 2021102042329; status of application: under application; specific aspect of manuscript covered in patent application: the gene panel predicting patients’ response to PARP inhibitor and cisplatin.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

