In-depth Temporal Transcriptome Profiling of Monkeypox and Host Cells using Nanopore Sequencing
- PMID: 37160911
- PMCID: PMC10170163
- DOI: 10.1038/s41597-023-02149-4
In-depth Temporal Transcriptome Profiling of Monkeypox and Host Cells using Nanopore Sequencing
Abstract
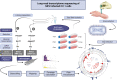
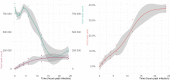
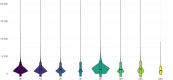
The recent human Monkeypox outbreak underlined the importance of studying basic biology of orthopoxviruses. However, the transcriptome of its causative agent has not been investigated before neither with short-, nor with long-read sequencing approaches. This Oxford Nanopore long-read RNA-Sequencing dataset fills this gap. It will enable the in-depth characterization of the transcriptomic architecture of the monkeypox virus, and may even make possible to annotate novel host transcripts. Moreover, our direct cDNA and native RNA sequencing reads will allow the estimation of gene expression changes of both the virus and the host cells during the infection. Overall, our study will lead to a deeper understanding of the alterations caused by the viral infection on a transcriptome level.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Exploring the transcriptomic profile of human monkeypox virus via CAGE and native RNA sequencing approaches.mSphere. 2024 Sep 25;9(9):e0035624. doi: 10.1128/msphere.00356-24. Epub 2024 Aug 27. mSphere. 2024. PMID: 39191390 Free PMC article.
-
High temporal resolution Nanopore sequencing dataset of SARS-CoV-2 and host cell RNAs.Gigascience. 2022 Oct 17;11:giac094. doi: 10.1093/gigascience/giac094. Gigascience. 2022. PMID: 36251275 Free PMC article.
-
Transcriptome profiling for precision cancer medicine using shallow nanopore cDNA sequencing.Sci Rep. 2023 Feb 9;13(1):2378. doi: 10.1038/s41598-023-29550-8. Sci Rep. 2023. PMID: 36759549 Free PMC article.
-
Long-Read Sequencing - A Powerful Tool in Viral Transcriptome Research.Trends Microbiol. 2019 Jul;27(7):578-592. doi: 10.1016/j.tim.2019.01.010. Epub 2019 Feb 26. Trends Microbiol. 2019. PMID: 30824172 Review.
-
Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data.Trends Genet. 2022 Mar;38(3):246-257. doi: 10.1016/j.tig.2021.09.001. Epub 2021 Oct 25. Trends Genet. 2022. PMID: 34711425 Review.
Cited by
-
Multi-omics characterization of the monkeypox virus infection.Nat Commun. 2024 Aug 8;15(1):6778. doi: 10.1038/s41467-024-51074-6. Nat Commun. 2024. PMID: 39117661 Free PMC article.
-
Transcriptomics data for muscle development in Goats.Sci Data. 2025 Jun 2;12(1):928. doi: 10.1038/s41597-025-04950-9. Sci Data. 2025. PMID: 40456801 Free PMC article.
-
Long-read Transcriptomics of Caviid Gammaherpesvirus 1: Compiling a Comprehensive RNA Atlas.bioRxiv [Preprint]. 2024 Dec 13:2024.12.11.627975. doi: 10.1101/2024.12.11.627975. bioRxiv. 2024. Update in: mSystems. 2025 Mar 18;10(3):e0167824. doi: 10.1128/msystems.01678-24. PMID: 39713410 Free PMC article. Updated. Preprint.
-
Long-read transcriptomics of caviid gammaherpesvirus 1: compiling a comprehensive RNA atlas.mSystems. 2025 Mar 18;10(3):e0167824. doi: 10.1128/msystems.01678-24. Epub 2025 Feb 27. mSystems. 2025. PMID: 40013795 Free PMC article.
-
Monkeypox virus genomic accordion strategies.Nat Commun. 2024 Apr 18;15(1):3059. doi: 10.1038/s41467-024-46949-7. Nat Commun. 2024. PMID: 38637500 Free PMC article.
References
-
- Moss, B. & Smith, G. L. Poxviridae: The Viruses and Their Replication. in Field’s Virology 573–613 (2021).
-
- Elwood JM. Smallpox and its eradication. J. Epidemiol. Community Heal. 1989;43:92–92. doi: 10.1136/jech.43.1.92. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

