In-depth Temporal Transcriptome Profiling of Monkeypox and Host Cells using Nanopore Sequencing
- PMID: 37160911
- PMCID: PMC10170163
- DOI: 10.1038/s41597-023-02149-4
In-depth Temporal Transcriptome Profiling of Monkeypox and Host Cells using Nanopore Sequencing
Abstract
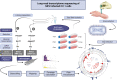
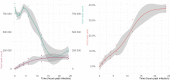
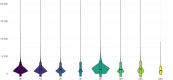
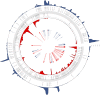
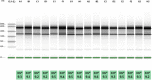
The recent human Monkeypox outbreak underlined the importance of studying basic biology of orthopoxviruses. However, the transcriptome of its causative agent has not been investigated before neither with short-, nor with long-read sequencing approaches. This Oxford Nanopore long-read RNA-Sequencing dataset fills this gap. It will enable the in-depth characterization of the transcriptomic architecture of the monkeypox virus, and may even make possible to annotate novel host transcripts. Moreover, our direct cDNA and native RNA sequencing reads will allow the estimation of gene expression changes of both the virus and the host cells during the infection. Overall, our study will lead to a deeper understanding of the alterations caused by the viral infection on a transcriptome level.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Moss, B. & Smith, G. L. Poxviridae: The Viruses and Their Replication. in Field’s Virology 573–613 (2021).
-
- Elwood JM. Smallpox and its eradication. J. Epidemiol. Community Heal. 1989;43:92–92. doi: 10.1136/jech.43.1.92. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

