Distinct oncogenic phenotypes in hematopoietic specific deletions of Trp53
- PMID: 37160922
- PMCID: PMC10169790
- DOI: 10.1038/s41598-023-33949-8
Distinct oncogenic phenotypes in hematopoietic specific deletions of Trp53
Abstract
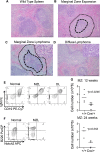
Loss of function in the tumor suppressor gene TP53 is the most common alteration seen in human cancer. In mice, P53 deletion in all cells leads predominantly to the development of T-cell lymphomas, followed by B-cell lymphomas, sarcomas and teratomas. In order to dissect the role of P53 in the hematopoietic system, we generated and analyzed two different mouse models deficient for P53. A pan-hematopoietic P53 deletion mouse was created using Vav1-Cre based deletion; and a B-cell-specific deletion mouse was created using a CD19-Cre based deletion. The Vav1-P53CKO mice predominantly developed T-cell malignancies in younger mice, and myeloid malignancies in older mice. In T-cell malignancies, there was accelerated thymic cell maturation with overexpression of Notch1 and its downstream effectors. CD19-P53CKO mice developed marginal zone expansion in the spleen, followed by marginal zone lymphoma, some of which progressed to diffuse large B-cell lymphomas. Interestingly, marginal zone and diffuse large B-cell lymphomas had a unique gene expression signature characterized by activation of the PI3K pathway, compared with wild type marginal zone or follicular cells of the spleen. This study demonstrates lineage specific P53 deletion leading to distinct phenotypes secondary to unique gene expression programs set in motion.
© 2023. The Author(s).
Conflict of interest statement
Dr. Rao has received compensation as a consultant for AbbVie, Inc. The remaining authors declare no relevant conflicts of interest.
Figures





References
-
- Levine AJ. The P53 tumor suppressor gene and gene product. Princess Takamatsu Symp. 1989;20:221–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32CA009120/NH/NIH HHS/United States
- T32 CA009056/NH/NIH HHS/United States
- R01CA264986/NH/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA264986/CA/NCI NIH HHS/United States
- R35 GM130361/GM/NIGMS NIH HHS/United States
- R01 CA166450/CA/NCI NIH HHS/United States
- P30 CA016042/CA/NCI NIH HHS/United States
- P30 AI028697/AI/NIAID NIH HHS/United States
- K08AR072787/NH/NIH HHS/United States
- T32 CA009120/CA/NCI NIH HHS/United States
- T32 CA009056/CA/NCI NIH HHS/United States
- R01 GM109146/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

