Double emulsions as a high-throughput enrichment and isolation platform for slower-growing microbes
- PMID: 37160952
- PMCID: PMC10169782
- DOI: 10.1038/s43705-023-00241-9
Double emulsions as a high-throughput enrichment and isolation platform for slower-growing microbes
Abstract
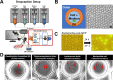
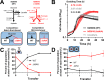
Our understanding of in situ microbial physiology is primarily based on physiological characterization of fast-growing and readily-isolatable microbes. Microbial enrichments to obtain novel isolates with slower growth rates or physiologies adapted to low nutrient environments are plagued by intrinsic biases for fastest-growing species when using standard laboratory isolation protocols. New cultivation tools to minimize these biases and enrich for less well-studied taxa are needed. In this study, we developed a high-throughput bacterial enrichment platform based on single cell encapsulation and growth within double emulsions (GrowMiDE). We showed that GrowMiDE can cultivate many different microorganisms and enrich for underrepresented taxa that are never observed in traditional batch enrichments. For example, preventing dominance of the enrichment by fast-growing microbes due to nutrient privatization within the double emulsion droplets allowed cultivation of slower-growing Negativicutes and Methanobacteria from stool samples in rich media enrichment cultures. In competition experiments between growth rate and growth yield specialist strains, GrowMiDE enrichments prevented competition for shared nutrient pools and enriched for slower-growing but more efficient strains. Finally, we demonstrated the compatibility of GrowMiDE with commercial fluorescence-activated cell sorting (FACS) to obtain isolates from GrowMiDE enrichments. Together, GrowMiDE + DE-FACS is a promising new high-throughput enrichment platform that can be easily applied to diverse microbial enrichments or screens.
© 2023. The Author(s).
Conflict of interest statement
Methods and techniques outlined in this work are disclosed in a U.S. patent filing, U.S. PTO Application No. 62/693800, filed by co-authors K. K. B, S. K., and P. M. F.
Figures


References
-
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;293:1436. - PubMed

