Weakly supervised detection and classification of basal cell carcinoma using graph-transformer on whole slide images
- PMID: 37160953
- PMCID: PMC10169852
- DOI: 10.1038/s41598-023-33863-z
Weakly supervised detection and classification of basal cell carcinoma using graph-transformer on whole slide images
Erratum in
-
Author Correction: Weakly supervised detection and classification of basal cell carcinoma using graph-transformer on whole slide images.Sci Rep. 2023 Aug 22;13(1):13678. doi: 10.1038/s41598-023-40875-2. Sci Rep. 2023. PMID: 37608213 Free PMC article. No abstract available.
Abstract
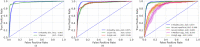
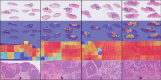
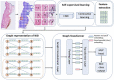
The high incidence rates of basal cell carcinoma (BCC) cause a significant burden at pathology laboratories. The standard diagnostic process is time-consuming and prone to inter-pathologist variability. Despite the application of deep learning approaches in grading of other cancer types, there is limited literature on the application of vision transformers to BCC on whole slide images (WSIs). A total of 1832 WSIs from 479 BCCs, divided into training and validation (1435 WSIs from 369 BCCs) and testing (397 WSIs from 110 BCCs) sets, were weakly annotated into four aggressivity subtypes. We used a combination of a graph neural network and vision transformer to (1) detect the presence of tumor (two classes), (2) classify the tumor into low and high-risk subtypes (three classes), and (3) classify four aggressivity subtypes (five classes). Using an ensemble model comprised of the models from cross-validation, accuracies of 93.5%, 86.4%, and 72% were achieved on two, three, and five class classifications, respectively. These results show high accuracy in both tumor detection and grading of BCCs. The use of automated WSI analysis could increase workflow efficiency.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

