Beyond assembly: the increasing flexibility of single-molecule sequencing technology
- PMID: 37161088
- PMCID: PMC10169143
- DOI: 10.1038/s41576-023-00600-1
Beyond assembly: the increasing flexibility of single-molecule sequencing technology
Abstract
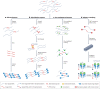
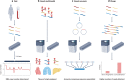
The maturation of high-throughput short-read sequencing technology over the past two decades has shaped the way genomes are studied. Recently, single-molecule, long-read sequencing has emerged as an essential tool in deciphering genome structure and function, including filling gaps in the human reference genome, measuring the epigenome and characterizing splicing variants in the transcriptome. With recent technological developments, these single-molecule technologies have moved beyond genome assembly and are being used in a variety of ways, including to selectively sequence specific loci with long reads, measure chromatin state and protein-DNA binding in order to investigate the dynamics of gene regulation, and rapidly determine copy number variation. These increasingly flexible uses of single-molecule technologies highlight a young and fast-moving part of the field that is leading to a more accessible era of nucleic acid sequencing.
© 2023. Springer Nature Limited.
Conflict of interest statement
W.T. has two patents (8,748,091 and 8,394,584) licensed to ONT. W.T. has received travel funds to speak at symposia organized by ONT. P.H. declares no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

