Neurologic sequelae of COVID-19 are determined by immunologic imprinting from previous coronaviruses
- PMID: 37161609
- PMCID: PMC11004923
- DOI: 10.1093/brain/awad155
Neurologic sequelae of COVID-19 are determined by immunologic imprinting from previous coronaviruses
Abstract
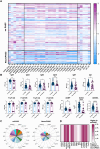
Coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains a global public health emergency. Although SARS-CoV-2 is primarily a respiratory pathogen, extra-respiratory organs, including the CNS, can also be affected. Neurologic symptoms have been observed not only during acute SARS-CoV-2 infection, but also at distance from respiratory disease, also known as long-COVID or neurological post-acute sequelae of COVID-19 (neuroPASC). The pathogenesis of neuroPASC is not well understood, but hypotheses include SARS-CoV-2-induced immune dysfunctions, hormonal dysregulations and persistence of SARS-CoV-2 reservoirs. In this prospective cohort study, we used a high throughput systems serology approach to dissect the humoral response to SARS-CoV-2 (and other common coronaviruses: 229E, HKU1, NL63 and OC43) in the serum and CSF from 112 infected individuals who developed (n = 18) or did not develop (n = 94) neuroPASC. Unique SARS-CoV-2 humoral profiles were observed in the CSF of neuroPASC compared with serum responses. All antibody isotypes (IgG, IgM, IgA) and subclasses (IgA1-2, IgG1-4) were detected in serum, whereas CSF was characterized by focused IgG1 (and absence of IgM). These data argue in favour of compartmentalized brain-specific responses against SARS-CoV-2 through selective transfer of antibodies from the serum to the CSF across the blood-brain barrier, rather than intrathecal synthesis, where more diversity in antibody classes/subclasses would be expected. Compared to individuals who did not develop post-acute complications following infection, individuals with neuroPASC had similar demographic features (median age 65 versus 66.5 years, respectively, P = 0.55; females 33% versus 44%, P = 0.52) but exhibited attenuated systemic antibody responses against SARS-CoV-2, characterized by decreased capacity to activate antibody-dependent complement deposition (ADCD), NK cell activation (ADNKA) and to bind Fcγ receptors. However, surprisingly, neuroPASC individuals showed significantly expanded antibody responses to other common coronaviruses, including 229E, HKU1, NL63 and OC43. This biased humoral activation across coronaviruses was particularly enriched in neuroPASC individuals with poor outcome, suggesting an 'original antigenic sin' (or immunologic imprinting), where pre-existing immune responses against related viruses shape the response to the current infection, as a key prognostic marker of neuroPASC disease. Overall, these findings point to a pathogenic role for compromised anti-SARS-CoV-2 responses in the CSF, likely resulting in incomplete virus clearance from the brain and persistent neuroinflammation, in the development of post-acute neurologic complications of SARS-CoV-2 infection.
Keywords: PASC; SARS-CoV-2; antibody-mediated complement activation; fc receptor; long-COVID.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
S.M. received speaker honoraria from Novartis and Biogen. G.A. is a founder of SeromYx Systems, Inc. and is a member of the scientific advisory board of Sanofi Pasteur. The other authors report no competing interests.
Figures




Comment in
-
Can immunological imprinting drive neurological dysfunction in long COVID?Brain. 2023 Oct 3;146(10):3960-3962. doi: 10.1093/brain/awad307. Brain. 2023. PMID: 37681523 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 AR072577/AR/NIAMS NIH HHS/United States
- UL1 TR002541/TR/NCATS NIH HHS/United States
- K23 AR073334/AR/NIAMS NIH HHS/United States
- P30 AR070253/AR/NIAMS NIH HHS/United States
- R01 AI042790/AI/NIAID NIH HHS/United States
- R01 AI146785/AI/NIAID NIH HHS/United States
- R01 NS122570/NS/NINDS NIH HHS/United States
- 75N93019C00052/AI/NIAID NIH HHS/United States
- R03 AR078938/AR/NIAMS NIH HHS/United States
- U19 AI135995/AI/NIAID NIH HHS/United States
- R01 AR077607/AR/NIAMS NIH HHS/United States
- U01 CA260476/CA/NCI NIH HHS/United States
- R37 AI080289/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

