Generation of Synthetic Acinetobacter baumannii-Specific Nanobodies
- PMID: 37162304
- PMCID: PMC10262196
- DOI: 10.1021/acsinfecdis.3c00024
Generation of Synthetic Acinetobacter baumannii-Specific Nanobodies
Abstract
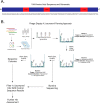
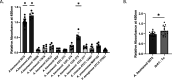
The bacterial pathogen Acinetobacter baumannii is a leading cause of drug-resistant infections. Here, we investigated the potential of developing nanobodies that can recognize A. baumannii over other Gram-negative bacteria. Through generation and panning of a synthetic nanobody library, we identified several potential lead candidates. We demonstrate how incorporation of next-generation sequencing analysis can aid in the selection of lead candidate nanobodies. Using monoclonal phage display, we validated the binding of lead nanobodies to A. baumannii. Subsequent purification and biochemical characterization revealed one particularly robust nanobody that specifically bound select A. baumannii strains compared to other common drug-resistant pathogens. These findings support the potential for nanobodies to selectively target A. baumannii and the identification of lead candidates for future investigation.
Keywords: Acinetobacter baumannii; drug resistance; nanobodies; phage display; sequencing.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Zollner-Schwetz I.; Zechner E.; Ullrich E.; Luxner J.; Pux C.; Pichler G.; Schippinger W.; Krause R.; Leitner E. Colonization of Long Term Care Facility Patients with MDR-Gram-Negatives during an Acinetobacter Baumannii Outbreak. Antimicrob. Resist. Infect. Control 2017, 6, 49.10.1186/s13756-017-0209-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

