This is a preprint.
TorsinA is essential for the timing and localization of neuronal nuclear pore complex biogenesis
- PMID: 37162852
- PMCID: PMC10168336
- DOI: 10.1101/2023.04.26.538491
TorsinA is essential for the timing and localization of neuronal nuclear pore complex biogenesis
Update in
-
TorsinA is essential for neuronal nuclear pore complex localization and maturation.Nat Cell Biol. 2024 Sep;26(9):1482-1495. doi: 10.1038/s41556-024-01480-1. Epub 2024 Aug 8. Nat Cell Biol. 2024. PMID: 39117796 Free PMC article.
Abstract
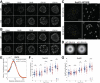
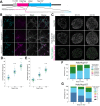
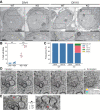
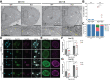
Nuclear pore complexes (NPCs) regulate information transfer between the nucleus and cytoplasm. NPC defects are linked to several neurological diseases, but the processes governing NPC biogenesis and spatial organization are poorly understood. Here, we identify a temporal window of strongly upregulated NPC biogenesis during neuronal maturation. We demonstrate that the AAA+ protein torsinA, whose loss of function causes the neurodevelopmental movement disorder DYT-TOR1A (DYT1) dystonia, coordinates NPC spatial organization during this period without impacting total NPC density. Using a new mouse line in which endogenous Nup107 is Halo-Tagged, we find that torsinA is essential for correct localization of NPC formation. In the absence of torsinA, the inner nuclear membrane buds excessively at sites of mislocalized, nascent NPCs, and NPC assembly completion is delayed. Our work implies that NPC spatial organization and number are independently regulated and suggests that torsinA is critical for the normal localization and assembly kinetics of NPCs.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
