This is a preprint.
Designer DNA NanoGripper
- PMID: 37162861
- PMCID: PMC10168355
- DOI: 10.1101/2023.04.26.538490
Designer DNA NanoGripper
Update in
-
Bioinspired designer DNA NanoGripper for virus sensing and potential inhibition.Sci Robot. 2024 Nov 27;9(96):eadi2084. doi: 10.1126/scirobotics.adi2084. Epub 2024 Nov 27. Sci Robot. 2024. PMID: 39602515
Abstract
DNA has shown great biocompatibility, programmable mechanical properties, and structural addressability at the nanometer scale, making it a versatile material for building high precision nanorobotics for biomedical applications. Herein, we present design principle, synthesis, and characterization of a DNA nanorobotic hand, called the "NanoGripper", that contains a palm and four bendable fingers as inspired by human hands, bird claws, and bacteriophages evolved in nature. Each NanoGripper finger has three phalanges connected by two flexible and rotatable joints that are bendable in response to binding to other entities. Functions of the NanoGripper have been enabled and driven by the interactions between moieties attached to the fingers and their binding partners. We showcase that the NanoGripper can be engineered to interact with and capture various objects with different dimensions, including gold nanoparticles, gold NanoUrchins, and SARS-CoV-2 virions. When carrying multiple DNA aptamer nanoswitches programmed to generate fluorescent signal enhanced on a photonic crystal platform, the NanoGripper functions as a sensitive viral biosensor that detects intact SARS-CoV-2 virions in human saliva with a limit of detection of ~ 100 copies/mL, providing RT-PCR equivalent sensitivity. Additionally, we use confocal microscopy to visualize how the NanoGripper-aptamer complex can effectively block viral entry into the host cells, indicating the viral inhibition. In summary, we report the design, synthesis, and characterization of a complex nanomachine that can be readily tailored for specific applications. The study highlights a path toward novel, feasible, and efficient solutions for the diagnosis and therapy of other diseases such as HIV and influenza.
Figures
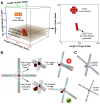

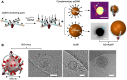




References
-
- Hu C., Pané S., Nelson B. J., Soft Micro- and Nanorobotics. Annual Review of Control, Robotics, and Autonomous Systems 1, 53–75 (2018).
-
- Wang B., Kostarelos K., Nelson B. J., Zhang L., Trends in Micro-/Nanorobotics: Materials Development, Actuation, Localization, and System Integration for Biomedical Applications. Advanced Materials 33, (2020). - PubMed
-
- Lafontaine D. L. J., Tollervey D., The function and synthesis of ribosomes. Nature Reviews Molecular Cell Biology 2, 514–520 (2001). - PubMed
-
- Hirokawa N., Noda Y., Tanaka Y., Niwa S., Kinesin superfamily motor proteins and intracellular transport. Nature Reviews Molecular Cell Biology 10, 682–696 (2009). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
