This is a preprint.
Competition between transcription and loop extrusion modulates promoter and enhancer dynamics
- PMID: 37162887
- PMCID: PMC10168261
- DOI: 10.1101/2023.04.25.538222
Competition between transcription and loop extrusion modulates promoter and enhancer dynamics
Update in
-
Transcription processes compete with loop extrusion to homogenize promoter and enhancer dynamics.Sci Adv. 2024 Dec 13;10(50):eadq0987. doi: 10.1126/sciadv.adq0987. Epub 2024 Dec 13. Sci Adv. 2024. PMID: 39671497 Free PMC article.
Abstract
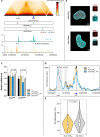
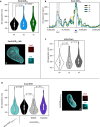
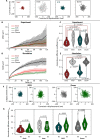
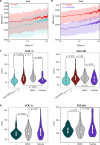
The spatiotemporal configuration of genes with distal regulatory elements, and the impact of chromatin mobility on transcription, remain unclear. Loop extrusion is an attractive model for bringing genetic elements together, but how this functionally interacts with transcription is also largely unknown. We combine live tracking of genomic loci and nascent transcripts with molecular dynamics simulations to assess the 4D arrangement of the Sox2 gene and its enhancer, in response to a battery of perturbations. We find that alterations in chromatin mobility, not promoter-enhancer distance, is more informative about transcriptional status. Active elements display more constrained mobility, consistent with confinement within specialized nuclear sites, and alterations in enhancer mobility distinguish poised from transcribing alleles. Strikingly, we find that whereas loop extrusion and transcription factor-mediated clustering contribute to promoter-enhancer proximity, they have antagonistic effects on chromatin dynamics. This provides an experimental framework for the underappreciated role of chromatin dynamics in genome regulation.
Keywords: Chromatin dynamics; anomalous diffusion; enhancer; live imaging; molecular dynamics; transcription.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures
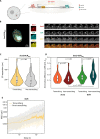
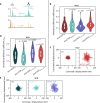

References
-
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, et al. (2016) Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–84. 10.1016/j.cell.2016.09.037. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
