This is a preprint.
Identification of FacZ as a division site placement factor in Staphylococcus aureus
- PMID: 37162900
- PMCID: PMC10168275
- DOI: 10.1101/2023.04.24.538170
Identification of FacZ as a division site placement factor in Staphylococcus aureus
Update in
-
FacZ is a GpsB-interacting protein that prevents aberrant division-site placement in Staphylococcus aureus.Nat Microbiol. 2024 Mar;9(3):801-813. doi: 10.1038/s41564-024-01607-y. Epub 2024 Mar 5. Nat Microbiol. 2024. PMID: 38443581 Free PMC article.
Abstract
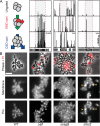
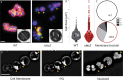
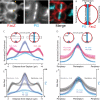
Staphylococcus aureus is a gram-positive pathogen responsible for life-threatening infections that are difficult to treat due to antibiotic resistance. The identification of new vulnerabilities in essential processes like cell envelope biogenesis represents a promising avenue towards the development of anti-staphylococcal therapies that overcome resistance. To this end, we performed cell sorting-based enrichments for S. aureus mutants with defects in envelope integrity and cell division. We identified many known envelope biogenesis factors as well as a large collection of new factors with roles in this process. Mutants inactivated for one of the hits, the uncharacterized SAOUHSC_01855 protein, displayed aberrant membrane invaginations and multiple FtsZ cytokinetic ring structures. This factor is broadly distributed among Firmicutes, and its inactivation in B. subtilis similarly caused division and membrane defects. We therefore renamed the protein FacZ (Firmicute-associated coordinator of Z-rings). In S. aureus, inactivation of the conserved cell division protein GpsB suppressed the division and morphological defects of facZ mutants. Additionally, FacZ and GpsB were found to interact directly in a purified system. Thus, FacZ is a novel antagonist of GpsB function with a conserved role in controlling division site placement in S. aureus and other Firmicutes.
Figures



References
-
- Koch A. L. Growth and Form of the Bacterial Cell Wall. American Scientist 78, 327–341 (1990).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
