This is a preprint.
The Genome-wide Signature of Short-term Temporal Selection
- PMID: 37162919
- PMCID: PMC10168312
- DOI: 10.1101/2023.04.28.538790
The Genome-wide Signature of Short-term Temporal Selection
Update in
-
The genome-wide signature of short-term temporal selection.Proc Natl Acad Sci U S A. 2024 Jul 9;121(28):e2307107121. doi: 10.1073/pnas.2307107121. Epub 2024 Jul 3. Proc Natl Acad Sci U S A. 2024. PMID: 38959040 Free PMC article.
Abstract
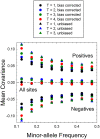
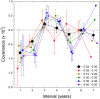
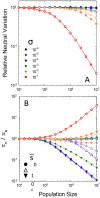
Despite evolutionary biology's obsession with natural selection, few studies have evaluated multi-generational series of patterns of selection on a genome-wide scale in natural populations. Here, we report on a nine-year population-genomic survey of the microcrustacean Daphnia pulex. The genome-sequences of > 800 isolates provide insights into patterns of selection that cannot be obtained from long-term molecular-evolution studies, including the pervasiveness of near quasi-neutrality across the genome (mean net selection coefficients near zero, but with significant temporal variance about the mean, and little evidence of positive covariance of selection across time intervals), the preponderance of weak negative selection operating on minor alleles, and a genome-wide distribution of numerous small linkage islands of observable selection influencing levels of nucleotide diversity. These results suggest that fluctuating selection is a major determinant of standing levels of variation in natural populations, challenge the conventional paradigm for interpreting patterns of nucleotide diversity and divergence, and motivate the need for the development of new theoretical expressions for the interpretation of population-genomic data.
Keywords: Daphnia pulex; effective population size; fluctuating selection; genetic diversity; linked selection effects; natural selection; population genomics; quasi-neutrality.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
