This is a preprint.
Detection of distant familial relatedness in biobanks for identification of undiagnosed carriers of a Mendelian disease variant: application to Long QT syndrome
- PMID: 37163006
- PMCID: PMC10168417
- DOI: 10.1101/2023.04.19.23288831
Detection of distant familial relatedness in biobanks for identification of undiagnosed carriers of a Mendelian disease variant: application to Long QT syndrome
Abstract
Importance: The diagnosis and study of rare genetic disease is often limited to referral populations, leading to underdiagnosis and a biased assessment of penetrance and phenotype.
Objective: To develop a generalizable method of genotype inference based on distant relatedness and to deploy this to identify undiagnosed Type 5 Long QT Syndrome (LQT5) rare variant carriers in a non-referral population.
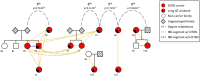
Participants: We identified 9 LQT5 probands and 3 first-degree relatives referred to a single Genetic Arrhythmia clinic, each carrying D76N (p.Asp76Asn), the most common variant implicated in LQT5. The non-referral population consisted of 69,879 ancestry-matched subjects in BioVU, a large biobank that links electronic health records to dense array data. Participants were enrolled from 2007-2022. Data analysis was performed in 2022.
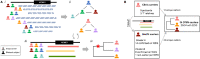
Exposures: We developed and applied a novel approach to genotype inference (Distant Relatedness for Identification and Variant Evaluation, or DRIVE) to identify shared, identical-by-descent (IBD) large chromosomal segments in array data.
Main outcomes and measures: We sought to establish genetic relatedness among the probands and to use genomic segments underlying D76N to identify other potential carriers in BioVU. We then further studied the role of D76N in LQT5 pathogenesis.
Results: Genetic reconstruction of pedigrees and distant relatedness detection among clinic probands using DRIVE revealed shared recent common ancestry and identified a single long shared haplotype. Interrogation of the non-referral population in BioVU identified a further 23 subjects sharing this haplotype, and sequencing confirmed D76N carrier status in 22, all previously undiagnosed with LQT5. The QTc was prolonged in D76N carriers compared to BioVU controls, with 40% penetrance of QTc ≥ 480 msec. Among D76N carriers, a QTc polygenic score was additively associated with QTc prolongation.
Conclusions and relevance: Detection of IBD shared chromosomal segments around D76N enabled identification of distantly related and previously undiagnosed rare-variant carriers, demonstrated the contribution of polygenic risk to monogenic disease penetrance, and further established LQT5 as a primary arrhythmia disorder. Analysis of shared chromosomal regions spanning disease-causing mutations can identify undiagnosed cases of genetic diseases.
Conflict of interest statement
Disclosures None of these activities are related to the content of this work. The other authors report no conflicts.
Figures


References
-
- Krahn AD, Laksman Z, Sy RW, et al. Congenital Long QT Syndrome. JACC Clin Electrophysiol. 2022;8(5):687–706. - PubMed
-
- Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet. 1997;17(3):338–340. - PubMed
-
- Sanguinetti MC, Curran ME, Zou A, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384(6604):80–83. - PubMed
-
- Yang T, Kupershmidt S, Roden DM. Anti-minK antisense decreases the amplitude of the rapidly activating cardiac delayed rectifier K+ current. Circ Res. 1995;77(6):1246–1253. - PubMed
-
- McDonald TV, Yu Z, Ming Z, et al. A minK-HERG complex regulates the cardiac potassium current I(Kr). Nature. 1997;388(6639):289–292. - PubMed
Publication types
Grants and funding
- S10 OD017985/OD/NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- T32 HG008341/HG/NHGRI NIH HHS/United States
- R01 GM133169/GM/NIGMS NIH HHS/United States
- U19 HL065962/HL/NHLBI NIH HHS/United States
- S10 RR025141/RR/NCRR NIH HHS/United States
- T32 HG008962/HG/NHGRI NIH HHS/United States
- R01 HD074711/HD/NICHD NIH HHS/United States
- RC2 GM092618/GM/NIGMS NIH HHS/United States
- P50 GM115305/GM/NIGMS NIH HHS/United States
- U01 HG011181/HG/NHGRI NIH HHS/United States
- S10 OD025092/OD/NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- R01 NS032830/NS/NINDS NIH HHS/United States
- U01 HG004798/HG/NHGRI NIH HHS/United States
- UL1 TR002243/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
