This is a preprint.
Single-cell profiling reveals immune aberrations in progressive idiopathic pulmonary fibrosis
- PMID: 37163015
- PMCID: PMC10168511
- DOI: 10.1101/2023.04.29.23289296
Single-cell profiling reveals immune aberrations in progressive idiopathic pulmonary fibrosis
Update in
-
Single-Cell Profiling Reveals Immune Aberrations in Progressive Idiopathic Pulmonary Fibrosis.Am J Respir Crit Care Med. 2024 Aug 15;210(4):484-496. doi: 10.1164/rccm.202306-0979OC. Am J Respir Crit Care Med. 2024. PMID: 38717443 Free PMC article.
Abstract
Rationale: Changes in peripheral blood cell populations have been observed but not detailed at single-cell resolution in idiopathic pulmonary fibrosis (IPF).
Objectives: To provide an atlas of the changes in the peripheral immune system in stable and progressive IPF.
Methods: Peripheral blood mononuclear cells (PBMCs) from IPF patients and controls were profiled using 10x Chromium 5' single-cell RNA sequencing (scRNA-seq). Flow cytometry was used for validation. Protein concentrations of Regulatory T-cells (Tregs) and Monocytes chemoattractants were measured in plasma and lung homogenates from patients and controls.
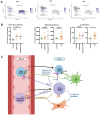
Measurements and main results: Thirty-eight PBMC samples from 25 patients with IPF and 13 matched controls yielded 149,564 cells that segregated into 23 subpopulations, corresponding to all expected peripheral blood cell populations. Classical monocytes were increased in progressive and stable IPF compared to controls (32.1%, 25.2%, 17.9%, respectively, p<0.05). Total lymphocytes were decreased in IPF vs controls, and in progressive vs stable IPF (52.6% vs 62.6%, p=0.035). Tregs were increased in progressive IPF (1.8% vs 1.1%, p=0.007), and were associated with decreased survival (P=0.009 in Kaplan-Meier analysis). Flow cytometry analysis confirmed this finding in an independent cohort of IPF patients. Tregs were also increased in two cohorts of lung scRNA-seq. CCL22 and CCL18, ligands for CCR4 and CCR8 Treg chemotaxis receptors, were increased in IPF.
Conclusions: The single-cell atlas of the peripheral immune system in IPF, reveals an outcome-predictive increase in classical monocytes and Tregs, as well as evidence for a lung-blood immune recruitment axis involving CCL7 (for classical monocytes) and CCL18/CCL22 (for Tregs).
Keywords: Idiopathic Pulmonary Fibrosis; Immune System; Monocytes; Regulatory T Cells; Single-cell RNA Sequencing.
Conflict of interest statement
NK is a scientific founder at Thyron, served as a consultant to Biogen Idec, Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Theravance, LifeMax, Three Lake Partners, Optikira, Astra Zeneca, RohBar, Veracyte, Augmanity, CSL Behring, Galapagos, Fibrogen, and Thyron over the last 3 years, reports Equity in Pliant and Thyron, and grants from Veracyte, Boehringer Ingelheim, BMS and non-financial support from MiRagen and Astra Zeneca. AU reports receiving research funding from Boehringer Ingelheim, and personal consulting fees or honoraria from Boehringer Ingelheim, Kamada, RemedyCell, Augmanity Nano, Splisense, Veracyte, and 1E Therapeutics in the last 36 months. JCS reports receiving honoraria and travel support from Boehringer Ingelheim. All other authors report no conflict of interest.
Figures





References
-
- Raghu G, Remy-Jardin M, Richeldi L, Thomson CC, Inoue Y, Johkoh T, Kreuter M, Lynch DA, Maher TM, Martinez FJ, Molina-Molina M, Myers JL, Nicholson AG, Ryerson CJ, Strek ME, Troy LK, Wijsenbeek M, Mammen MJ, Hossain T, Bissell BD, Herman DD, Hon SM, Kheir F, Khor YH, Macrea M, Antoniou KM, Bouros D, Buendia-Roldan I, Caro F, Crestani B, Ho L, Morisset J, Olson AL, Podolanczuk A, Poletti V, Selman M, Ewing T, Jones S, Knight SL, Ghazipura M, Wilson KC. Idiopathic Pulmonary Fibrosis (an Update) and Progressive Pulmonary Fibrosis in Adults: An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med 2022; 205: e18–e47. - PMC - PubMed
-
- Kolahian S, Fernandez IE, Eickelberg O, Haiti D. Immune Mechanisms in Pulmonary Fibrosis. Am J Respir Cell Mol Biol 2016; 55: 309–322. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
