This is a preprint.
A transposase-derived gene required for human brain development
- PMID: 37163102
- PMCID: PMC10168387
- DOI: 10.1101/2023.04.28.538770
A transposase-derived gene required for human brain development
Abstract
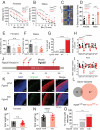
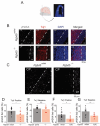
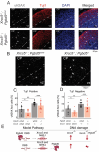
DNA transposable elements and transposase-derived genes are present in most living organisms, including vertebrates, but their function is largely unknown. PiggyBac Transposable Element Derived 5 (PGBD5) is an evolutionarily conserved vertebrate DNA transposase-derived gene with retained nuclease activity in human cells. Vertebrate brain development is known to be associated with prominent neuronal cell death and DNA breaks, but their causes and functions are not well understood. Here, we show that PGBD5 contributes to normal brain development in mice and humans, where its deficiency causes disorder of intellectual disability, movement, and seizures. In mice, Pgbd5 is required for the developmental induction of post-mitotic DNA breaks and recurrent somatic genome rearrangements. In the brain cortex, loss of Pgbd5 leads to aberrant differentiation and gene expression of distinct neuronal populations, including specific types of glutamatergic neurons, which explains the features of PGBD5 deficiency in humans. Thus, PGBD5 might be a transposase-derived enzyme required for brain development in mammals.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests. AK is a consultant for Novartis, Rgenta, Blueprint, and Syndax. RR is a founder and a member of the SAB of Genotwin, and a member of the SAB of Diatech Pharmacogenetics. None of these activities are related to the work described in this manuscript.
Figures



References
-
- Edelman G. M., Neural Darwinism: selection and reentrant signaling in higher brain function. Neuron 10, 115–125 (1993). - PubMed
-
- Barnes D. E., Stamp G., Rosewell I., Denzel A., Lindahl T., Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Current biology : CB 8, 1395–1398 (1998). - PubMed
-
- Gao Y., Chaudhuri J., Zhu C., Davidson L., Weaver D. T., Alt F. W., A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 9, 367–376 (1998). - PubMed
-
- Frank K. M. et al., Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396, 173–177 (1998). - PubMed
-
- Sekiguchi J. M. et al., Nonhomologous end-joining proteins are required for V(D)J recombination, normal growth, and neurogenesis. Cold Spring Harb Symp Quant Biol 64, 169–181 (1999). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
