This is a preprint.
Bcl-xL is translocated to the nucleus via CtBP2 to epigenetically promote metastasis
- PMID: 37163116
- PMCID: PMC10168309
- DOI: 10.1101/2023.04.26.538373
Bcl-xL is translocated to the nucleus via CtBP2 to epigenetically promote metastasis
Update in
-
Bcl-xL is translocated to the nucleus via CtBP2 to epigenetically promote metastasis.Cancer Lett. 2024 Nov 1;604:217240. doi: 10.1016/j.canlet.2024.217240. Epub 2024 Sep 10. Cancer Lett. 2024. PMID: 39265800 Free PMC article.
Abstract
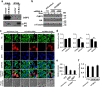
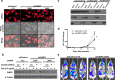
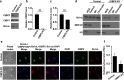
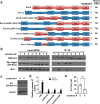
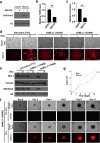
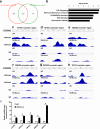
Besides its mitochondria-based anti-apoptotic role, Bcl-xL also travels to the nucleus to promote cancer metastasis by upregulating global histone H3 trimethyl Lys4 (H3K4me3) and TGFβ transcription. How Bcl-xL is translocated into the nucleus and how nuclear Bcl-xL regulates H3K4me3 modification are not understood. Here, we report that C-terminal Binding Protein 2 (CtBP2) binds Bcl-xL via its N-terminus and translocates Bcl-xL into the nucleus. Knockdown of CtBP2 by shRNA decreases the nuclear portion of Bcl-xL and reverses Bcl-xL-induced cell migration and metastasis in mouse models. Furthermore, knockout of CtBP2 suppresses Bcl-xL transcription. The binding between Bcl-xL and CtBP2 is required for their interaction with MLL1, a histone H3K4 methyltransferase. Pharmacologic inhibition of MLL1 enzymatic activity reverses Bcl-xL-induced H3K4me3 and TGFβ mRNA upregulation as well as cell invasion. Moreover, cleavage under targets and release using nuclease (CUT&RUN) coupled with next generation sequencing reveals that H3K4me3 modifications are particularly enriched in the promotor region of genes encoding TGFβ and its signaling pathway in the cancer cells overexpressing Bcl-xL. Altogether, the metastatic function of Bcl-xL is mediated by its interaction with CtBP2 and MLL1.
Keywords: Bcl-xL; CtBP2; MLL1; epigenetic modification; metastasis.
Conflict of interest statement
Competing interests The authors declare no competing financial interests.
Figures


References
-
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB (1993) bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74: 597–608 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
