Centralspindlin proteins Pavarotti and Tumbleweed along with WASH regulate nuclear envelope budding
- PMID: 37163553
- PMCID: PMC10174194
- DOI: 10.1083/jcb.202211074
Centralspindlin proteins Pavarotti and Tumbleweed along with WASH regulate nuclear envelope budding
Abstract
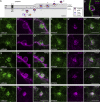
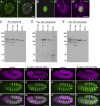
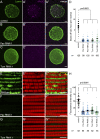
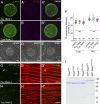
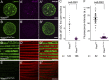
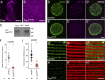
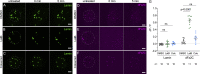
Nuclear envelope (NE) budding is a nuclear pore-independent nuclear export pathway, analogous to the egress of herpesviruses, and required for protein quality control, synapse development, and mitochondrial integrity. The physical formation of NE buds is dependent on the Wiskott-Aldrich Syndrome protein, Wash, its regulatory complex (SHRC), and Arp2/3, and requires Wash's actin nucleation activity. However, the machinery governing cargo recruitment and organization within the NE bud remains unknown. Here, we identify Pavarotti (Pav) and Tumbleweed (Tum) as new molecular components of NE budding. Pav and Tum interact directly with Wash and define a second nuclear Wash-containing complex required for NE budding. Interestingly, we find that the actin-bundling activity of Pav is required, suggesting a structural role in the physical and/or organizational aspects of NE buds. Thus, Pav and Tum are providing exciting new entry points into the physical machineries of this alternative nuclear export pathway for large cargos during cell differentiation and development.
© 2023 Davidson et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

