Butt-seq: a new method for facile profiling of transcription
- PMID: 37164645
- PMCID: PMC10270195
- DOI: 10.1101/gad.350434.123
Butt-seq: a new method for facile profiling of transcription
Abstract
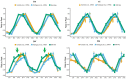
A wide range of sequencing methods has been developed to assess nascent RNA transcription and resolve the single-nucleotide position of RNA polymerase genome-wide. These techniques are often burdened with high input material requirements and lengthy protocols. We leveraged the template-switching properties of thermostable group II intron reverse transcriptase (TGIRT) and developed Butt-seq (bulk analysis of nascent transcript termini sequencing), which can produce libraries from purified nascent RNA in 6 h and from as few as 10,000 cells-an improvement of at least 10-fold over existing techniques. Butt-seq shows that inhibition of the superelongation complex (SEC) causes promoter-proximal pausing to move upstream in a fashion correlated with subnucleosomal fragments. To address transcriptional regulation in a tissue, Butt-seq was used to measure the circadian regulation of transcription from fly heads. All the results indicate that Butt-seq is a simple and powerful technique to analyze transcription at a high level of resolution.
Keywords: RNA polymerase II pausing; circadian rhythms; nascent RNA; superelongation complex; transcriptional profiling; transcriptional regulation.
© 2023 Yu and Rosbash; Published by Cold Spring Harbor Laboratory Press.
Figures







References
-
- Begik O, Diensthuber G, Liu H, Delgado-Tejedor A, Kontur C, Niazi AM, Valen E, Giraldez AJ, Beaudoin JD, Mattick JS, et al. 2023. Nano3P-seq: transcriptome-wide analysis of gene expression and tail dynamics using end-capture nanopore cDNA sequencing. Nat Methods 20: 75–85. 10.1038/s41592-022-01714-w - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
