Mitotic clustering of pulverized chromosomes from micronuclei
- PMID: 37165191
- PMCID: PMC10307639
- DOI: 10.1038/s41586-023-05974-0
Mitotic clustering of pulverized chromosomes from micronuclei
Abstract
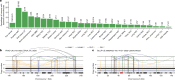
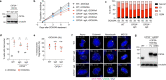
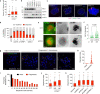
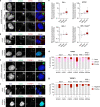
Complex genome rearrangements can be generated by the catastrophic pulverization of missegregated chromosomes trapped within micronuclei through a process known as chromothripsis1-5. As each chromosome contains a single centromere, it remains unclear how acentric fragments derived from shattered chromosomes are inherited between daughter cells during mitosis6. Here we tracked micronucleated chromosomes with live-cell imaging and show that acentric fragments cluster in close spatial proximity throughout mitosis for asymmetric inheritance by a single daughter cell. Mechanistically, the CIP2A-TOPBP1 complex prematurely associates with DNA lesions within ruptured micronuclei during interphase, which poises pulverized chromosomes for clustering upon mitotic entry. Inactivation of CIP2A-TOPBP1 caused acentric fragments to disperse throughout the mitotic cytoplasm, stochastically partition into the nucleus of both daughter cells and aberrantly misaccumulate as cytoplasmic DNA. Mitotic clustering facilitates the reassembly of acentric fragments into rearranged chromosomes lacking the extensive DNA copy-number losses that are characteristic of canonical chromothripsis. Comprehensive analysis of pan-cancer genomes revealed clusters of DNA copy-number-neutral rearrangements-termed balanced chromothripsis-across diverse tumour types resulting in the acquisition of known cancer driver events. Thus, distinct patterns of chromothripsis can be explained by the spatial clustering of pulverized chromosomes from micronuclei.
© 2023. The Author(s).
Conflict of interest statement
S.F.B. owns equity in, receives compensation from and serves as a consultant and on the scientific advisory board and board of directors of Volastra Therapeutics. The other authors declare no competing interests.
Figures
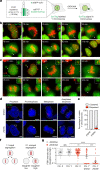
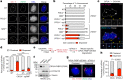
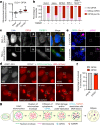
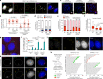








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

