NAD+ metabolism-based immunoregulation and therapeutic potential
- PMID: 37165408
- PMCID: PMC10171153
- DOI: 10.1186/s13578-023-01031-5
NAD+ metabolism-based immunoregulation and therapeutic potential
Abstract
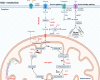
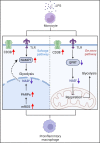
Nicotinamide adenine dinucleotide (NAD+) is a critical metabolite that acts as a cofactor in energy metabolism, and serves as a cosubstrate for non-redox NAD+-dependent enzymes, including sirtuins, CD38 and poly(ADP-ribose) polymerases. NAD+ metabolism can regulate functionality attributes of innate and adaptive immune cells and contribute to inflammatory responses. Thus, the manipulation of NAD+ bioavailability can reshape the courses of immunological diseases. Here, we review the basics of NAD+ biochemistry and its roles in the immune response, and discuss current challenges and the future translational potential of NAD+ research in the development of therapeutics for inflammatory diseases, such as COVID-19.
Keywords: Disease therapy; Immunoregulation; Metabolic homeostasis; Nicotinamide adenine dinucleotide (NAD+); Plasticity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

