Two interacting ethylene response factors negatively regulate peach resistance to Lasiodiplodia theobromae
- PMID: 37165714
- PMCID: PMC10400035
- DOI: 10.1093/plphys/kiad279
Two interacting ethylene response factors negatively regulate peach resistance to Lasiodiplodia theobromae
Abstract
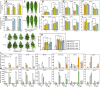
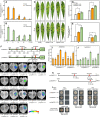
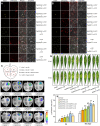
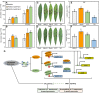
Gummosis is 1 of the most common and destructive diseases threatening global peach (Prunus persica) production. Our previous studies have revealed that ethylene and methyl jasmonate enhance peach susceptibility to Lasiodiplodia theobromae, a virulent pathogen inducing gummosis; however, the underlying molecular mechanisms remain obscure. Here, 2 ethylene response factors (ERFs), PpERF98 and PpERF1, were identified as negative regulators in peach response to L. theobromae infection. Expression of 2 putative paralogs, PpERF98-1/2, was dramatically induced by ethylene and L. theobromae treatments and accumulated highly in the gummosis-sensitive cultivar. Silencing of PpERF98-1/2 increased salicylic acid (SA) content and pathogenesis-related genes PpPR1 and PpPR2 transcripts, conferring peach resistance to L. theobromae, whereas peach and tomato (Solanum lycopersicum) plants overexpressing either of PpERF98-1/2 showed opposite changes. Also, jasmonic acid markedly accumulated in PpERF98-1/2-silenced plants, but reduction in PpPR3, PpPR4, and PpCHI (Chitinase) transcripts indicated a blocked signaling pathway. PpERF98-1 and 2 were further demonstrated to directly bind the promoters of 2 putative paralogous PpERF1 genes and to activate the ERF branch of the jasmonate/ethylene signaling pathway, thus attenuating SA-dependent defenses. The lesion phenotypes of peach seedlings overexpressing PpERF1-1/2 and PpERF98-1/2 were similar. Furthermore, PpERF98-1/2 formed homodimers/heterodimers and interacted with the 2 PpERF1 proteins to amplify the jasmonate/ethylene signaling pathway, as larger lesions were observed in peach plants cooverexpressing PpERF98 with PpERF1 relative to individual PpERF98 overexpression. Overall, our work deciphers an important regulatory network of ethylene-mediated peach susceptibility to L. theobromae based on a PpERF98-PpERF1 transcriptional cascade, which could be utilized as a potential target for genetic engineering to augment protection against L. theobromae-mediated diseases in crops and trees.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



Similar articles
-
Lasiodiplodia theobromae-induced alteration in ROS metabolism and its relation to gummosis development in Prunus persica.Plant Physiol Biochem. 2020 Sep;154:43-53. doi: 10.1016/j.plaphy.2020.05.018. Epub 2020 May 22. Plant Physiol Biochem. 2020. PMID: 32526610
-
Integrated transcriptomic and metabolic analyses reveal that ethylene enhances peach susceptibility to Lasiodiplodia theobromae-induced gummosis.Hortic Res. 2022 Jan 5;9:uhab019. doi: 10.1093/hr/uhab019. Hortic Res. 2022. PMID: 35040976 Free PMC article.
-
Gene Expression Changes during the Gummosis Development of Peach Shoots in Response to Lasiodiplodia theobromae Infection Using RNA-Seq.Front Physiol. 2016 May 9;7:170. doi: 10.3389/fphys.2016.00170. eCollection 2016. Front Physiol. 2016. PMID: 27242544 Free PMC article.
-
Characterization of phytotoxin and secreted proteins identifies of Lasiodiplodia theobromae, causes of peach gummosis.Fungal Biol. 2019 Jan;123(1):51-58. doi: 10.1016/j.funbio.2018.11.001. Epub 2018 Nov 14. Fungal Biol. 2019. PMID: 30654957
-
Silicon inhibits gummosis in peach via ethylene and PpERF-PpPG1 pathway.Plant Sci. 2022 Sep;322:111362. doi: 10.1016/j.plantsci.2022.111362. Epub 2022 Jun 23. Plant Sci. 2022. PMID: 35753620
Cited by
-
Characterization of metallothionein genes from Broussonetia papyrifera: metal binding and heavy metal tolerance mechanisms.BMC Genomics. 2024 Jun 5;25(1):563. doi: 10.1186/s12864-024-10477-x. BMC Genomics. 2024. PMID: 38840042 Free PMC article.
-
A comparative study of Machine Learning-based classification of Tomato fungal diseases: Application of GLCM texture features.Heliyon. 2023 Oct 31;9(11):e21697. doi: 10.1016/j.heliyon.2023.e21697. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027996 Free PMC article.
-
Genome-wide analysis of AP2/ERF gene family and functional characterization of StoERF109 in Solanum torvum response to Verticillium dahliae infection.Planta. 2025 May 20;262(1):1. doi: 10.1007/s00425-025-04723-z. Planta. 2025. PMID: 40392396
-
Functional analysis of PagERF021 gene in salt stress tolerance in Populus alba × P. glandulosa.Plant Genome. 2024 Dec;17(4):e20521. doi: 10.1002/tpg2.20521. Epub 2024 Oct 16. Plant Genome. 2024. PMID: 39414577 Free PMC article.
-
ZmNLR-7-Mediated Synergistic Regulation of ROS, Hormonal Signaling, and Defense Gene Networks Drives Maize Immunity to Southern Corn Leaf Blight.Curr Issues Mol Biol. 2025 Jul 21;47(7):573. doi: 10.3390/cimb47070573. Curr Issues Mol Biol. 2025. PMID: 40729042 Free PMC article.
References
-
- Achuo EA, Audenaert K, Meziane H, Hofte M. The salicylic acid-dependent defence pathway is effective against different pathogens in tomato and tobacco. Plant Pathol. 2004:53(1):65–72. 10.1111/j.1365-3059.2004.00947.x - DOI
-
- Beckman TG, Pusey PL, Bertrand PF. Impact of fungal gummosis on peach trees. HortScience 2003:38(6):1141–1143. 10.21273/HORTSCI.38.6.1141 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

