Epigenotype-genotype-phenotype correlations in SETD1A and SETD2 chromatin disorders
- PMID: 37166351
- PMCID: PMC10630252
- DOI: 10.1093/hmg/ddad079
Epigenotype-genotype-phenotype correlations in SETD1A and SETD2 chromatin disorders
Abstract
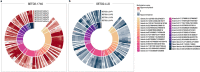
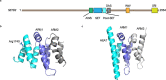
Germline pathogenic variants in two genes encoding the lysine-specific histone methyltransferase genes SETD1A and SETD2 are associated with neurodevelopmental disorders (NDDs) characterized by developmental delay and congenital anomalies. The SETD1A and SETD2 gene products play a critical role in chromatin-mediated regulation of gene expression. Specific methylation episignatures have been detected for a range of chromatin gene-related NDDs and have impacted clinical practice by improving the interpretation of variant pathogenicity. To investigate if SETD1A and/or SETD2-related NDDs are associated with a detectable episignature, we undertook targeted genome-wide methylation profiling of > 2 M CpGs using a next-generation sequencing-based assay. A comparison of methylation profiles in patients with SETD1A variants (n = 6) did not reveal evidence of a strong methylation episignature. A review of the clinical and genetic features of the SETD2 patient group revealed that, as reported previously, there were phenotypic differences between patients with truncating mutations (n = 4, Luscan-Lumish syndrome; MIM:616831) and those with missense codon 1740 variants [p.Arg1740Trp (n = 4) and p.Arg1740Gln (n = 2)]. Both SETD2 subgroups demonstrated a methylation episignature, which was characterized by hypomethylation and hypermethylation events, respectively. Within the codon 1740 subgroup, both the methylation changes and clinical phenotype were more severe in those with p.Arg1740Trp variants. We also noted that two of 10 cases with a SETD2-NDD had developed a neoplasm. These findings reveal novel epigenotype-genotype-phenotype correlations in SETD2-NDDs and predict a gain-of-function mechanism for SETD2 codon 1740 pathogenic variants.
© The Author(s) 2023. Published by Oxford University Press.
Figures




References
-
- Aref-Eshghi, E., Rodenhiser, D.I., Schenkel, L.C., Lin, H., Skinner, C., Ainsworth, P., Paré, G., Hood, R.L., Bulman, D.E., Kernohan, K.D.et al. (2018) Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am. J. Hum. Genet., 102, 156–174. - PMC - PubMed
-
- Levy, M.A., Relator, R., McConkey, H., Pranckeviciene, E., Kerkhof, J., Barat-Houari, M., Bargiacchi, S., Biamino, E., Palomares Bralo, M., Cappuccio, G.et al. (2022) Functional correlation of genome-wide DNA methylation profiles in genetic neurodevelopmental disorders. Hum. Mutat., 43, 1609–1628. - PubMed

