Mitoribosome Biogenesis
- PMID: 37166630
- PMCID: PMC10639111
- DOI: 10.1007/978-1-0716-3171-3_3
Mitoribosome Biogenesis
Abstract
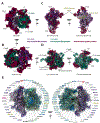
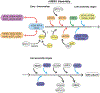
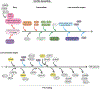
Mitoribosome biogenesis is a complex and energetically costly process that involves RNA elements encoded in the mitochondrial genome and mitoribosomal proteins most frequently encoded in the nuclear genome. The process is catalyzed by extra-ribosomal proteins, nucleus-encoded assembly factors that act in all stages of the assembly process to coordinate the processing and maturation of ribosomal RNAs with the hierarchical association of ribosomal proteins. Biochemical studies and recent cryo-EM structures of mammalian mitoribosomes have provided hints regarding their assembly. In this general concept chapter, we will briefly describe the current knowledge, mainly regarding the mammalian mitoribosome biogenesis pathway and factors involved, and will emphasize the biological sources and approaches that have been applied to advance the field.
Keywords: Mitochondrial disease; Mitochondrial ribosome; Mitochondrial translation; Mitoribosome assembly; OXPHOS deficiency.
© 2023. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
None declared.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

