Identification of potentially pathogenic variants for autism spectrum disorders using gene-burden analysis
- PMID: 37167322
- PMCID: PMC10174571
- DOI: 10.1371/journal.pone.0273957
Identification of potentially pathogenic variants for autism spectrum disorders using gene-burden analysis
Abstract
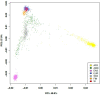
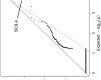
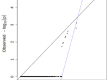
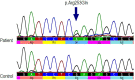
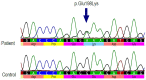
Gene- burden analyses have lately become a very successful way for the identification of genes carrying risk variants underlying the analysed disease. This approach is also suitable for complex disorders like autism spectrum disorder (ASD). The gene-burden analysis using Testing Rare Variants with Public Data (TRAPD) software was conducted on whole exome sequencing data of Slovenian patients with ASD to determine potentially novel disease risk variants in known ASD-associated genes as well as in others. To choose the right control group for testing, principal component analysis based on the 1000 Genomes and ASD cohort samples was conducted. The subsequent protein structure and ligand binding analysis usingI-TASSER package were performed to detect changes in protein structure and ligand binding to determine a potential pathogenic consequence of observed mutation. The obtained results demonstrate an association of two variants-p.Glu198Lys (PPP2R5D:c.592G>A) and p.Arg253Gln (PPP2R5D:c.758G>A) with the ASD. Substitution p.Glu198Lys (PPP2R5D:c.592G>A) is a variant, previously described as pathogenic in association with ASD combined with intellectual disability, whereas p.Arg253Gln (PPP2R5D:c.758G>A) has not been described as an ASD-associated pathogenic variant yet. The results indicate that the filtering process was suitable and could be used in the future for detection of novel pathogenic variants when analysing groups of ASD patients.
Copyright: © 2023 Rihar et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
NO. The authors have declared that no competing interests exist.
Figures



References
-
- Association AP. Diagnostic and statistical manual of mental disorders (DSM-5®): American Psychiatric Pub; 2013. - PubMed

