A genomic enhancer signature associates with hepatocellular carcinoma prognosis
- PMID: 37168287
- PMCID: PMC10165154
- DOI: 10.1016/j.jhepr.2023.100715
A genomic enhancer signature associates with hepatocellular carcinoma prognosis
Abstract
Background & aims: Lifestyle and environmental-related exposures are important risk factors for hepatocellular carcinoma (HCC), suggesting that epigenetic dysregulation significantly underpins HCC. We profiled 30 surgically resected tumours and the matched adjacent normal tissues to understand the aberrant epigenetic events associated with HCC.
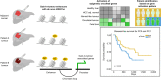
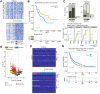
Methods: We identified tumour differential enhancers and the associated genes by analysing H3K27 acetylation (H3K27ac) chromatin immunoprecipitation sequencing (ChIP-seq) and Hi-C/HiChIP data from the resected tumour samples of 30 patients with early-stage HCC. This epigenome dataset was analysed with previously reported genome and transcriptome data of the overlapping group of patients from the same cohort. We performed patient-specific differential expression testing using multiregion sequencing data to identify genes that undergo both enhancer and gene expression changes. Based on the genes selected, we identified two patient groups and performed a recurrence-free survival analysis.
Results: We observed large-scale changes in the enhancer distribution between HCC tumours and the adjacent normal samples. Many of the gain-in-tumour enhancers showed corresponding upregulation of the associated genes and vice versa, but much of the enhancer and gene expression changes were patient-specific. A subset of the upregulated genes was activated in a subgroup of patients' tumours. Recurrence-free survival analysis revealed that the patients with a more robust upregulation of those genes showed a worse prognosis.
Conclusions: We report the genomic enhancer signature associated with differential prognosis in HCC. Findings that cohere with oncofoetal reprogramming in HCC were underpinned by genome-wide enhancer rewiring. Our results present the epigenetic changes in HCC that offer the rational selection of epigenetic-driven gene targets for therapeutic intervention or disease prognostication in HCC.
Impact and implications: Lifestyle and environmental-related exposures are the important risk factors of hepatocellular carcinoma (HCC), suggesting that tumour-associated epigenetic dysregulations may significantly underpin HCC. We profiled tumour tissues and their matched normal from 30 patients with early-stage HCC to study the dysregulated epigenetic changes associated with HCC. By also analysing the patients' RNA-seq and clinical data, we found the signature genes - with epigenetic and transcriptomic dysregulation - associated with worse prognosis. Our findings suggest that systemic approaches are needed to consider the surrounding cellular environmental and epigenetic changes in HCC tumours.
Keywords: Cancer prognosis; Epigenetics; Hepatocellular carcinoma; Multi-omics; Personalised medicine.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures





References
-
- Martínez-Jiménez F., Muiños F., Sentís I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C., et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–572. - PubMed
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- London W.T., McGlynn K.A. 3rd ed. Oxford University Press; New York: 2006. Liver cancer. Cancer epidemiology and prevention.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

