Designing rice panicle architecture via developmental regulatory genes
- PMID: 37168816
- PMCID: PMC10165343
- DOI: 10.1270/jsbbs.22075
Designing rice panicle architecture via developmental regulatory genes
Abstract
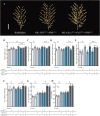
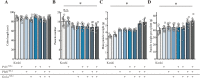
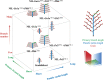
Rice panicle architecture displays remarkable diversity in branch number, branch length, and grain arrangement; however, much remains unknown about how such diversity in patterns is generated. Although several genes related to panicle branch number and panicle length have been identified, how panicle branch number and panicle length are coordinately regulated is unclear. Here, we show that panicle length and panicle branch number are independently regulated by the genes Prl5/OsGA20ox4, Pbl6/APO1, and Gn1a/OsCKX2. We produced near-isogenic lines (NILs) in the Koshihikari genetic background harboring the elite alleles for Prl5, regulating panicle rachis length; Pbl6, regulating primary branch length; and Gn1a, regulating panicle branching in various combinations. A pyramiding line carrying Prl5, Pbl6, and Gn1a showed increased panicle length and branching without any trade-off relationship between branch length or number. We successfully produced various arrangement patterns of grains by changing the combination of alleles at these three loci. Improvement of panicle architecture raised yield without associated negative effects on yield-related traits except for panicle number. Three-dimensional (3D) analyses by X-ray computed tomography (CT) of panicles revealed that differences in panicle architecture affect grain filling. Importantly, we determined that Prl5 improves grain filling without affecting grain number.
Keywords: grain filling; near-isogenic lines; panicle architecture; panicle development; rice.
Copyright © 2023 by JAPANESE SOCIETY OF BREEDING.
Figures



References
-
- Ashikari, M., Sakakibara H., Lin S.Y., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H. and Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745. - PubMed
-
- Ashikari, M. and Matsuoka M. (2006) Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci 11: 344–350. - PubMed

