Analysis of candidate genes for cleft lip ± cleft palate using murine single-cell expression data
- PMID: 37169019
- PMCID: PMC10165499
- DOI: 10.3389/fcell.2023.1091666
Analysis of candidate genes for cleft lip ± cleft palate using murine single-cell expression data
Abstract
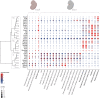
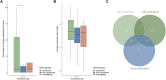
Introduction: Cleft lip ± cleft palate (CL/P) is one of the most common birth defects. Although research has identified multiple genetic risk loci for different types of CL/P (i.e., syndromic or non-syndromic forms), determining the respective causal genes and understanding the relevant functional networks remain challenging. The recent introduction of single-cell RNA sequencing (scRNA-seq) has provided novel opportunities to study gene expression patterns at cellular resolution. The aims of our study were to: (i) aggregate available scRNA-seq data from embryonic mice and provide this as a resource for the craniofacial community; and (ii) demonstrate the value of these data in terms of the investigation of the gene expression patterns of CL/P candidate genes. Methods and Results: First, two published scRNA-seq data sets from embryonic mice were re-processed, i.e., data representing the murine time period of craniofacial development: (i) facial data from embryonic day (E) E11.5; and (ii) whole embryo data from E9.5-E13.5 from the Mouse Organogenesis Cell Atlas (MOCA). Marker gene expression analyses demonstrated that at E11.5, the facial data were a high-resolution representation of the MOCA data. Using CL/P candidate gene lists, distinct groups of genes with specific expression patterns were identified. Among others we identified that a co-expression network including Irf6, Grhl3 and Tfap2a in the periderm, while it was limited to Irf6 and Tfap2a in palatal epithelia, cells of the ectodermal surface, and basal cells at the fusion zone. The analyses also demonstrated that additional CL/P candidate genes (e.g., Tpm1, Arid3b, Ctnnd1, and Wnt3) were exclusively expressed in Irf6+ facial epithelial cells (i.e., as opposed to Irf6- epithelial cells). The MOCA data set was finally used to investigate differences in expression profiles for candidate genes underlying different types of CL/P. These analyses showed that syndromic CL/P genes (syCL/P) were expressed in significantly more cell types than non-syndromic CL/P candidate genes (nsCL/P). Discussion: The present study illustrates how scRNA-seq data can empower research on craniofacial development and disease.
Keywords: IRF6; cleft lip with or without cleft palate; craniofacial development; expression pattern; single-cell RNA sequencing (scRNA-seq); single-cell transcriptomics.
Copyright © 2023 Siewert, Reiz, Krug, Heggemann, Mangold, Dickten and Ludwig.
Conflict of interest statement
BR and HD were employed by FASTGenomics (Comma Soft AG). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

