Degradomic Identification of Membrane Type 1-Matrix Metalloproteinase as an ADAMTS9 and ADAMTS20 Substrate
- PMID: 37169079
- PMCID: PMC10267602
- DOI: 10.1016/j.mcpro.2023.100566
Degradomic Identification of Membrane Type 1-Matrix Metalloproteinase as an ADAMTS9 and ADAMTS20 Substrate
Abstract
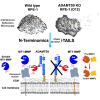
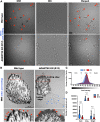
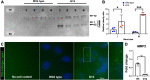
The secreted metalloproteases ADAMTS9 and ADAMTS20 are implicated in extracellular matrix proteolysis and primary cilium biogenesis. Here, we show that clonal gene-edited RPE-1 cells in which ADAMTS9 was inactivated, and which constitutively lack ADAMTS20 expression, have morphologic characteristics distinct from parental RPE-1 cells. To investigate underlying proteolytic mechanisms, a quantitative terminomics method, terminal amine isotopic labeling of substrates was used to compare the parental and gene-edited RPE-1 cells and their medium to identify ADAMTS9 substrates. Among differentially abundant neo-amino (N) terminal peptides arising from secreted and transmembrane proteins, a peptide with lower abundance in the medium of gene-edited cells suggested cleavage at the Tyr314-Gly315 bond in the ectodomain of the transmembrane metalloprotease membrane type 1-matrix metalloproteinase (MT1-MMP), whose mRNA was also reduced in gene-edited cells. This cleavage, occurring in the MT1-MMP hinge, that is, between the catalytic and hemopexin domains, was orthogonally validated both by lack of an MT1-MMP catalytic domain fragment in the medium of gene-edited cells and restoration of its release from the cell surface by reexpression of ADAMTS9 and ADAMTS20 and was dependent on hinge O-glycosylation. A C-terminally semitryptic MT1-MMP peptide with greater abundance in WT RPE-1 medium identified a second ADAMTS9 cleavage site in the MT1-MMP hemopexin domain. Consistent with greater retention of MT1-MMP on the surface of gene-edited cells, pro-MMP2 activation, which requires cell surface MT1-MMP, was increased. MT1-MMP knockdown in gene-edited ADAMTS9/20-deficient cells restored focal adhesions but not ciliogenesis. The findings expand the web of interacting proteases at the cell surface, suggest a role for ADAMTS9 and ADAMTS20 in regulating cell surface activity of MT1-MMP, and indicate that MT1-MMP shedding does not underlie their observed requirement in ciliogenesis.
Keywords: ADAM; ADAMTS; MMP; MT1-MMP; cleavage; degradome; degradomics; focal adhesion; gene-editing-N-terminomics; metalloprotease; protease; proteolysis; substrate.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures






References
-
- Kuno K., Matsushima K. ADAMTS-1 protein anchors at the extracellular matrix through the thrombospondin type I motifs and its spacing region. J. Biol. Chem. 1998;273:13912–13917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

