Efficient and accurate KIR and HLA genotyping with massively parallel sequencing data
- PMID: 37169596
- PMCID: PMC10519407
- DOI: 10.1101/gr.277585.122
Efficient and accurate KIR and HLA genotyping with massively parallel sequencing data
Abstract
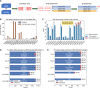
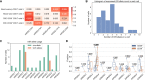
Killer cell immunoglobulin like receptor (KIR) genes and human leukocyte antigen (HLA) genes play important roles in innate and adaptive immunity. They are highly polymorphic and cannot be genotyped with standard variant calling pipelines. Compared with HLA genes, many KIR genes are similar to each other in sequences and may be absent in the chromosomes. Therefore, although many tools have been developed to genotype HLA genes using common sequencing data, none of them work for KIR genes. Even specialized KIR genotypers could not resolve all the KIR genes. Here we describe T1K, a novel computational method for the efficient and accurate inference of KIR or HLA alleles from RNA-seq, whole-genome sequencing, or whole-exome sequencing data. T1K jointly considers alleles across all genotyped genes, so it can reliably identify present genes and distinguish homologous genes, including the challenging KIR2DL5A/KIR2DL5B genes. This model also benefits HLA genotyping, where T1K achieves high accuracy in benchmarks. Moreover, T1K can call novel single-nucleotide variants and process single-cell data. Applying T1K to tumor single-cell RNA-seq data, we found that KIR2DL4 expression was enriched in tumor-specific CD8+ T cells. T1K may open the opportunity for HLA and KIR genotyping across various sequencing applications.
© 2023 Song et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Bhatt RS, Berjis A, Konge JC, Mahoney KM, Klee AN, Freeman SS, Chen C-H, Jegede OA, Catalano PJ, Pignon J-C, et al. 2021. KIR3DL3 is an inhibitory receptor for HHLA2 that mediates an alternative immunoinhibitory pathway to PD1. Cancer Immunol Res 9: 156–169. 10.1158/2326-6066.CIR-20-0315 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
