Clonal evolution during metastatic spread in high-risk neuroblastoma
- PMID: 37169874
- PMCID: PMC11481711
- DOI: 10.1038/s41588-023-01395-x
Clonal evolution during metastatic spread in high-risk neuroblastoma
Erratum in
-
Author Correction: Clonal evolution during metastatic spread in high-risk neuroblastoma.Nat Genet. 2025 Sep;57(9):2338. doi: 10.1038/s41588-025-02320-0. Nat Genet. 2025. PMID: 40764844 No abstract available.
Abstract
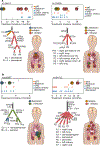
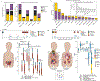
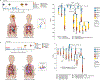
Patients with high-risk neuroblastoma generally present with widely metastatic disease and often relapse despite intensive therapy. As most studies to date focused on diagnosis-relapse pairs, our understanding of the genetic and clonal dynamics of metastatic spread and disease progression remain limited. Here, using genomic profiling of 470 sequential and spatially separated samples from 283 patients, we characterize subtype-specific genetic evolutionary trajectories from diagnosis through progression and end-stage metastatic disease. Clonal tracing timed disease initiation to embryogenesis. Continuous acquisition of structural variants at disease-defining loci (MYCN, TERT, MDM2-CDK4) followed by convergent evolution of mutations targeting shared pathways emerged as the predominant feature of progression. At diagnosis metastatic clones were already established at distant sites where they could stay dormant, only to cause relapses years later and spread via metastasis-to-metastasis and polyclonal seeding after therapy.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests Statement
G.G. is a consultant in Isabl Inc. E.P., A.L.K. and J.S.M-M are founders, equity holders and hold fiduciary roles in Isabl Inc. N-K.V.C reports receiving commercial research grants unrelated to this study, from Y-mabs Therapeutics and Abpro-Labs Inc.; holding ownership interest/equity in Y-Mabs Therapeutics Inc., holding ownership interest/equity in Abpro-Labs, and owning stock options in Eureka Therapeutics. N-K.V.C. is the inventor and owner of issued patents, some licensed by MSK to Ymabs Therapeutics, Biotec Pharmacon, and Abpro-labs. N-K.V.C. is an advisory board member for Abpro-Labs and Eureka Therapeutics. MSK also has financial interest in Y-mabs. The remaining authors declare no competing interests.
Figures








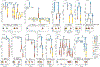






References
-
- Schramm A et al. Mutational dynamics between primary and relapse neuroblastomas. Nat. Genet 47, 872–877 (2015). - PubMed
Methods-only references
-
- Chakravarty D et al. OncoKB: Annotation of the oncogenic effect and treatment implications of somatic mutations in cancer. Journal of Clinical Oncology vol. 34 11583–11583 Preprint at 10.1200/jco.2016.34.15_suppl.11583 (2016). - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

