Widespread extinctions of co-diversified primate gut bacterial symbionts from humans
- PMID: 37169918
- PMCID: PMC10860671
- DOI: 10.1038/s41564-023-01388-w
Widespread extinctions of co-diversified primate gut bacterial symbionts from humans
Abstract
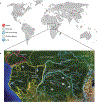
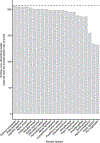
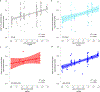
Humans and other primates harbour complex gut bacterial communities that influence health and disease, but the evolutionary histories of these symbioses remain unclear. This is partly due to limited information about the microbiota of ancestral primates. Here, using phylogenetic analyses of metagenome-assembled genomes (MAGs), we show that hundreds of gut bacterial clades diversified in parallel (that is, co-diversified) with primate species over millions of years, but that humans have experienced widespread losses of these ancestral symbionts. Analyses of 9,460 human and non-human primate MAGs, including newly generated MAGs from chimpanzees and bonobos, revealed significant co-diversification within ten gut bacterial phyla, including Firmicutes, Actinobacteriota and Bacteroidota. Strikingly, ~44% of the co-diversifying clades detected in African apes were absent from available metagenomic data from humans and ~54% were absent from industrialized human populations. In contrast, only ~3% of non-co-diversifying clades detected in African apes were absent from humans. Co-diversifying clades present in both humans and chimpanzees displayed consistent genomic signatures of natural selection between the two host species but differed in functional content from co-diversifying clades lost from humans, consistent with selection against certain functions. This study discovers host-species-specific bacterial symbionts that predate hominid diversification, many of which have undergone accelerated extinctions from human populations.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures


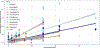



References
-
- Bello MG, Knight R, Gilbert JA & Blaser MJ Preserving microbial diversity. Science 362, 33–34 (2018). - PubMed
-
- Sonnenburg ED & Sonnenburg JL The ancestral and industrialised gut microbiota and implications for human health. Nat. Rev. Microbiol 17, 383–390 (2019). - PubMed
-
- Groussin M, Mazel F & Alm EJ Co-evolution and co-speciation of host-gut bacteria systems. Cell Host Microbe 28, 12–22 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

