The Swiss Pathogen Surveillance Platform - towards a nation-wide One Health data exchange platform for bacterial, viral and fungal genomics and associated metadata
- PMID: 37171846
- PMCID: PMC10272868
- DOI: 10.1099/mgen.0.001001
The Swiss Pathogen Surveillance Platform - towards a nation-wide One Health data exchange platform for bacterial, viral and fungal genomics and associated metadata
Abstract
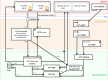
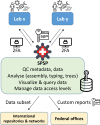
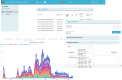
The Swiss Pathogen Surveillance Platform (SPSP) is a shared secure surveillance platform between human and veterinary medicine, to also include environmental and foodborne isolates. It enables rapid and detailed transmission monitoring and outbreak surveillance of pathogens using whole genome sequencing data and associated metadata. It features controlled data access, complex dynamic queries, dedicated dashboards and automated data sharing with international repositories, providing actionable results for public health and the vision to improve societal well-being and health.
Keywords: antibiotic resistance; bacteria; bioinformatics; database; epidemiology; fungi; molecular surveillance; outbreaks; research; sequencing; typing; virulence; virus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Improving the quality and workflow of bacterial genome sequencing and analysis: paving the way for a Switzerland-wide molecular epidemiological surveillance platform.Swiss Med Wkly. 2018 Dec 15;148:w14693. doi: 10.4414/smw.2018.14693. eCollection 2018 Dec 3. Swiss Med Wkly. 2018. PMID: 30552858
-
A Schema for Digitized Surface Swab Site Metadata in Open-Source DNA Sequence Databases.mSystems. 2023 Apr 27;8(2):e0128422. doi: 10.1128/msystems.01284-22. Epub 2023 Feb 27. mSystems. 2023. PMID: 36847566 Free PMC article.
-
Status and potential of bacterial genomics for public health practice: a scoping review.Implement Sci. 2019 Aug 13;14(1):79. doi: 10.1186/s13012-019-0930-2. Implement Sci. 2019. PMID: 31409417 Free PMC article.
-
ReporTree: a surveillance-oriented tool to strengthen the linkage between pathogen genetic clusters and epidemiological data.Genome Med. 2023 Jun 15;15(1):43. doi: 10.1186/s13073-023-01196-1. Genome Med. 2023. PMID: 37322495 Free PMC article.
-
Food Safety Genomics and Connections to One Health and the Clinical Microbiology Laboratory.Clin Lab Med. 2020 Dec;40(4):553-563. doi: 10.1016/j.cll.2020.08.011. Epub 2020 Oct 5. Clin Lab Med. 2020. PMID: 33121622 Review.
Cited by
-
Towards unified reporting of genome sequencing results in clinical microbiology.PeerJ. 2024 Aug 8;12:e17673. doi: 10.7717/peerj.17673. eCollection 2024. PeerJ. 2024. PMID: 39131622 Free PMC article.
-
The integrated genomic surveillance system of Andalusia (SIEGA) provides a One Health regional resource connected with the clinic.Sci Rep. 2024 Aug 19;14(1):19200. doi: 10.1038/s41598-024-70107-0. Sci Rep. 2024. PMID: 39160186 Free PMC article.
-
FAIR+E pathogen data for surveillance and research: lessons from COVID-19.Front Public Health. 2023 Nov 21;11:1289945. doi: 10.3389/fpubh.2023.1289945. eCollection 2023. Front Public Health. 2023. PMID: 38074768 Free PMC article.
-
IVDR: Analysis of the Social, Economic, and Practical Consequences of the Application of an Ordinance of the In Vitro Diagnostic Ordinance in Switzerland.Diagnostics (Basel). 2023 Sep 11;13(18):2910. doi: 10.3390/diagnostics13182910. Diagnostics (Basel). 2023. PMID: 37761277 Free PMC article.
-
Key role of whole genome sequencing in resolving an international outbreak of monophasic Salmonella Typhimurium linked to chocolate products.BMC Infect Dis. 2025 Feb 20;25(1):242. doi: 10.1186/s12879-025-10629-8. BMC Infect Dis. 2025. PMID: 39979856 Free PMC article.
References
-
- Lytsy B, Engstrand L, Gustafsson Å, Kaden R. Time to review the gold standard for genotyping vancomycin-resistant enterococci in epidemiology: comparing whole-genome sequencing with PFGE and MLST in three suspected outbreaks in Sweden during 2013-2015. Infect Genet Evol. 2017;54:74–80. doi: 10.1016/j.meegid.2017.06.010. - DOI - PubMed

