The molecular mechanism for carbon catabolite repression of the chitin response in Vibrio cholerae
- PMID: 37172034
- PMCID: PMC10208484
- DOI: 10.1371/journal.pgen.1010767
The molecular mechanism for carbon catabolite repression of the chitin response in Vibrio cholerae
Abstract
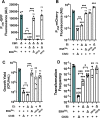
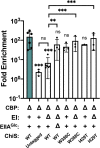
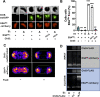
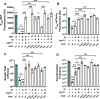
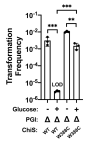
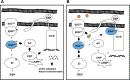
Vibrio cholerae is a facultative pathogen that primarily occupies marine environments. In this niche, V. cholerae commonly interacts with the chitinous shells of crustacean zooplankton. As a chitinolytic microbe, V. cholerae degrades insoluble chitin into soluble oligosaccharides. Chitin oligosaccharides serve as both a nutrient source and an environmental cue that induces a strong transcriptional response in V. cholerae. Namely, these oligosaccharides induce the chitin sensor, ChiS, to activate the genes required for chitin utilization and horizontal gene transfer by natural transformation. Thus, interactions with chitin impact the survival of V. cholerae in marine environments. Chitin is a complex carbon source for V. cholerae to degrade and consume, and the presence of more energetically favorable carbon sources can inhibit chitin utilization. This phenomenon, known as carbon catabolite repression (CCR), is mediated by the glucose-specific Enzyme IIA (EIIAGlc) of the phosphoenolpyruvate-dependent phosphotransferase system (PTS). In the presence of glucose, EIIAGlc becomes dephosphorylated, which inhibits ChiS transcriptional activity by an unknown mechanism. Here, we show that dephosphorylated EIIAGlc interacts with ChiS. We also isolate ChiS suppressor mutants that evade EIIAGlc-dependent repression and demonstrate that these alleles no longer interact with EIIAGlc. These findings suggest that EIIAGlc must interact with ChiS to exert its repressive effect. Importantly, the ChiS suppressor mutations we isolated also relieve repression of chitin utilization and natural transformation by EIIAGlc, suggesting that CCR of these behaviors is primarily regulated through ChiS. Together, our results reveal how nutrient conditions impact the fitness of an important human pathogen in its environmental reservoir.
Copyright: © 2023 Green et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Glucose-Specific Enzyme IIA of the Phosphoenolpyruvate:Carbohydrate Phosphotransferase System Modulates Chitin Signaling Pathways in Vibrio cholerae.J Bacteriol. 2017 Aug 22;199(18):e00127-17. doi: 10.1128/JB.00127-17. Print 2017 Sep 15. J Bacteriol. 2017. PMID: 28461445 Free PMC article.
-
Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression.Environ Microbiol. 2012 Aug;14(8):1898-912. doi: 10.1111/j.1462-2920.2011.02689.x. Epub 2012 Jan 6. Environ Microbiol. 2012. PMID: 22222000
-
Chitin-induced T6SS in Vibrio cholerae is dependent on ChiS activation.Microbiology (Reading). 2018 May;164(5):751-763. doi: 10.1099/mic.0.000656. Epub 2018 Apr 10. Microbiology (Reading). 2018. PMID: 29633936
-
Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae.Curr Opin Microbiol. 2016 Apr;30:1-7. doi: 10.1016/j.mib.2015.10.007. Epub 2015 Nov 23. Curr Opin Microbiol. 2016. PMID: 26615332 Review.
-
Chitin and Products of Its Hydrolysis in Vibrio cholerae Ecology.Biochemistry (Mosc). 2015 Sep;80(9):1109-16. doi: 10.1134/S0006297915090023. Biochemistry (Mosc). 2015. PMID: 26555464 Review.
Cited by
-
Two transmembrane transcriptional regulators coordinate to activate chitin-induced natural transformation in Vibrio cholerae.PLoS Genet. 2025 Feb 18;21(2):e1011606. doi: 10.1371/journal.pgen.1011606. eCollection 2025 Feb. PLoS Genet. 2025. PMID: 39965000 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

