Decoding murine cytomegalovirus
- PMID: 37172056
- PMCID: PMC10208470
- DOI: 10.1371/journal.ppat.1010992
Decoding murine cytomegalovirus
Abstract
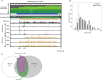
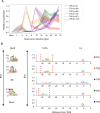
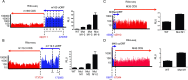
The genomes of both human cytomegalovirus (HCMV) and murine cytomegalovirus (MCMV) were first sequenced over 20 years ago. Similar to HCMV, the MCMV genome had initially been proposed to harbor ≈170 open reading frames (ORFs). More recently, omics approaches revealed HCMV gene expression to be substantially more complex comprising several hundred viral ORFs. Here, we provide a state-of-the art reannotation of lytic MCMV gene expression based on integrative analysis of a large set of omics data. Our data reveal 365 viral transcription start sites (TiSS) that give rise to 380 and 454 viral transcripts and ORFs, respectively. The latter include >200 small ORFs, some of which represented the most highly expressed viral gene products. By combining TiSS profiling with metabolic RNA labelling and chemical nucleotide conversion sequencing (dSLAM-seq), we provide a detailed picture of the expression kinetics of viral transcription. This not only resulted in the identification of a novel MCMV immediate early transcript encoding the m166.5 ORF, which we termed ie4, but also revealed a group of well-expressed viral transcripts that are induced later than canonical true late genes and contain an initiator element (Inr) but no TATA- or TATT-box in their core promoters. We show that viral upstream ORFs (uORFs) tune gene expression of longer viral ORFs expressed in cis at translational level. Finally, we identify a truncated isoform of the viral NK-cell immune evasin m145 arising from a viral TiSS downstream of the canonical m145 mRNA. Despite being ≈5-fold more abundantly expressed than the canonical m145 protein it was not required for downregulating the NK cell ligand, MULT-I. In summary, our work will pave the way for future mechanistic studies on previously unknown cytomegalovirus gene products in an important virus animal model.
Copyright: © 2023 Lodha et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Griffiths P, Reeves M. Pathogenesis of human cytomegalovirus in the immunocompromised host. Nat Rev Microbiol. 2021;19(12):759–73. Epub 2021/06/26. doi: 10.1038/s41579-021-00582-z ; PubMed Central PMCID: PMC8223196 number 2020135.6 assigned to University College London (UCL), entitled ‘hCMV antibody and vaccine target’, that deals with a novel antigenic domain on HCMV glycoprotein B (gB). UCL received funds from Takeda pharmaceuticals to compensate for the time P.G. spent as a member of the end-point committee for a randomized clinical trial (RCT) of maribavir. The authors declare no other competing interests. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

