Extracellular vesicles in fatty liver promote a metastatic tumor microenvironment
- PMID: 37172577
- PMCID: PMC10524732
- DOI: 10.1016/j.cmet.2023.04.013
Extracellular vesicles in fatty liver promote a metastatic tumor microenvironment
Abstract
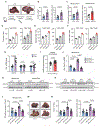
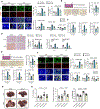
Liver metastasis is a major cause of death in patients with colorectal cancer (CRC). Fatty liver promotes liver metastasis, but the underlying mechanism remains unclear. We demonstrated that hepatocyte-derived extracellular vesicles (EVs) in fatty liver enhanced the progression of CRC liver metastasis by promoting oncogenic Yes-associated protein (YAP) signaling and an immunosuppressive microenvironment. Fatty liver upregulated Rab27a expression, which facilitated EV production from hepatocytes. In the liver, these EVs transferred YAP signaling-regulating microRNAs to cancer cells to augment YAP activity by suppressing LATS2. Increased YAP activity in CRC liver metastasis with fatty liver promoted cancer cell growth and an immunosuppressive microenvironment by M2 macrophage infiltration through CYR61 production. Patients with CRC liver metastasis and fatty liver had elevated nuclear YAP expression, CYR61 expression, and M2 macrophage infiltration. Our data indicate that fatty liver-induced EV-microRNAs, YAP signaling, and an immunosuppressive microenvironment promote the growth of CRC liver metastasis.
Keywords: CYR61; M2 macrophage; Rab27a; colon cancer; exosome; high-fat diet; liver metastasis; microRNA; non-alcoholic fatty liver disease; palmitate.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.N. is a member of the advisory board of Altimmune, BI, BMS, Cytodyn, 89BIO, EchoSens, Gilead, GSK, Madrgial, Merck, Novo Nordisk, OWL, Prespecturm, Pfizer, Roche Diagnostic, and Siemens, Terns and Takeda; received research support from Allergan, Akero, BMS, Gilead, Galectin, Genfit, GSK, Conatus, Corcept, Enanta, Madrigal, Novartis, Novo Nordisk, Shire, Takeda, Terns, Viking, and Zydus; and is a stockholder in Anaetos, Rivus Pharma, CIMA, ChronWell, and Viking.
Figures





Comment in
-
'Yapping' with extracellular vesicles in fatty liver metastasis.Trends Cell Biol. 2023 Sep;33(9):729-731. doi: 10.1016/j.tcb.2023.07.001. Epub 2023 Jul 18. Trends Cell Biol. 2023. PMID: 37468354

