Consensus clustering methodology to improve molecular stratification of non-small cell lung cancer
- PMID: 37173325
- PMCID: PMC10182023
- DOI: 10.1038/s41598-023-33954-x
Consensus clustering methodology to improve molecular stratification of non-small cell lung cancer
Abstract
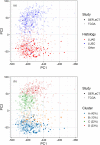
Recent advances in machine learning research, combined with the reduced sequencing costs enabled by modern next-generation sequencing, paved the way to the implementation of precision medicine through routine multi-omics molecular profiling of tumours. Thus, there is an emerging need of reliable models exploiting such data to retrieve clinically useful information. Here, we introduce an original consensus clustering approach, overcoming the intrinsic instability of common clustering methods based on molecular data. This approach is applied to the case of non-small cell lung cancer (NSCLC), integrating data of an ongoing clinical study (PROMOLE) with those made available by The Cancer Genome Atlas, to define a molecular-based stratification of the patients beyond, but still preserving, histological subtyping. The resulting subgroups are biologically characterized by well-defined mutational and gene-expression profiles and are significantly related to disease-free survival (DFS). Interestingly, it was observed that (1) cluster B, characterized by a short DFS, is enriched in KEAP1 and SKP2 mutations, that makes it an ideal candidate for further studies with inhibitors, and (2) over- and under-representation of inflammation and immune systems pathways in squamous-cell carcinomas subgroups could be potentially exploited to stratify patients treated with immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Ferlay, J. et al. Global cancer observatory: cancer today. Available at: https://gco.iarc.fr/today [Accessed 29/04/21].
-
- American Cancer Society. Cancer facts and figures. 2022:28.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

