Development and validation of a radiopathomic model for predicting pathologic complete response to neoadjuvant chemotherapy in breast cancer patients
- PMID: 37173635
- PMCID: PMC10176880
- DOI: 10.1186/s12885-023-10817-2
Development and validation of a radiopathomic model for predicting pathologic complete response to neoadjuvant chemotherapy in breast cancer patients
Abstract
Background: Neoadjuvant chemotherapy (NAC) has become the standard therapeutic option for early high-risk and locally advanced breast cancer. However, response rates to NAC vary between patients, causing delays in treatment and affecting the prognosis for patients who do not sensitive to NAC.
Materials and methods: In total, 211 breast cancer patients who completed NAC (training set: 155, validation set: 56) were retrospectively enrolled. we developed a deep learning radiopathomics model(DLRPM) by Support Vector Machine (SVM) method based on clinicopathological features, radiomics features, and pathomics features. Furthermore, we comprehensively validated the DLRPM and compared it with three single-scale signatures.
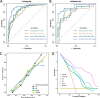
Results: DLRPM had favourable performance for the prediction of pathological complete response (pCR) in the training set (AUC 0.933[95% CI 0.895-0.971]), and in the validation set (AUC 0.927 [95% CI 0.858-0.996]). In the validation set, DLRPM also significantly outperformed the radiomics signature (AUC 0.821[0.700-0.942]), pathomics signature (AUC 0.766[0.629-0.903]), and deep learning pathomics signature (AUC 0.804[0.683-0.925]) (all p < 0.05). The calibration curves and decision curve analysis also indicated the clinical effectiveness of the DLRPM.
Conclusions: DLRPM can help clinicians accurately predict the efficacy of NAC before treatment, highlighting the potential of artificial intelligence to improve the personalized treatment of breast cancer patients.
Keywords: Breast cancer; CECT; Pathomics; Radiomics; Radiopathomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Development of a neoadjuvant chemotherapy efficacy prediction model for nasopharyngeal carcinoma integrating magnetic resonance radiomics and pathomics: a multi-center retrospective study.BMC Cancer. 2024 Dec 5;24(1):1501. doi: 10.1186/s12885-024-13235-0. BMC Cancer. 2024. PMID: 39639211 Free PMC article.
-
Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study.Lancet Digit Health. 2022 Jan;4(1):e8-e17. doi: 10.1016/S2589-7500(21)00215-6. Lancet Digit Health. 2022. PMID: 34952679
-
Deep learning radiomics of ultrasonography for comprehensively predicting tumor and axillary lymph node status after neoadjuvant chemotherapy in breast cancer patients: A multicenter study.Cancer. 2023 Feb 1;129(3):356-366. doi: 10.1002/cncr.34540. Epub 2022 Nov 19. Cancer. 2023. PMID: 36401611
-
Machine learning with magnetic resonance imaging for prediction of response to neoadjuvant chemotherapy in breast cancer: A systematic review and meta-analysis.Eur J Radiol. 2022 May;150:110247. doi: 10.1016/j.ejrad.2022.110247. Epub 2022 Mar 10. Eur J Radiol. 2022. PMID: 35290910
-
Advancing personalized oncology: a systematic review on the integration of artificial intelligence in monitoring neoadjuvant treatment for breast cancer patients.BMC Cancer. 2024 Oct 21;24(1):1300. doi: 10.1186/s12885-024-13049-0. BMC Cancer. 2024. PMID: 39434042 Free PMC article.
Cited by
-
Development and validation of a pathological model predicting the efficacy of neoadjuvant therapy for breast cancer based on RCB scoring.Arch Med Sci. 2024 May 1;21(1):92-101. doi: 10.5114/aoms/188006. eCollection 2025. Arch Med Sci. 2024. PMID: 40190325 Free PMC article.
-
An integrated approach of feature selection and machine learning for early detection of breast cancer.Sci Rep. 2025 Apr 15;15(1):13015. doi: 10.1038/s41598-025-97685-x. Sci Rep. 2025. PMID: 40234520 Free PMC article.
-
Personalized predictions of neoadjuvant chemotherapy response in breast cancer using machine learning and full-field digital mammography radiomics.Front Med (Lausanne). 2025 Apr 17;12:1582560. doi: 10.3389/fmed.2025.1582560. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40313551 Free PMC article.
-
Machine learning prediction of pathological complete response and overall survival of breast cancer patients in an underserved inner-city population.Breast Cancer Res. 2024 Jan 10;26(1):7. doi: 10.1186/s13058-023-01762-w. Breast Cancer Res. 2024. PMID: 38200586 Free PMC article.
-
Development of a neoadjuvant chemotherapy efficacy prediction model for nasopharyngeal carcinoma integrating magnetic resonance radiomics and pathomics: a multi-center retrospective study.BMC Cancer. 2024 Dec 5;24(1):1501. doi: 10.1186/s12885-024-13235-0. BMC Cancer. 2024. PMID: 39639211 Free PMC article.
References
-
- Guarneri V, Griguolo G, Miglietta F, Conte PF, Dieci MV, Girardi F. Survival after neoadjuvant therapy with trastuzumab-lapatinib and chemotherapy in patients with HER2-positive early breast cancer: a meta-analysis of randomized trials. ESMO Open. 2022;7(2):100433. doi: 10.1016/j.esmoop.2022.100433. - DOI - PMC - PubMed
-
- Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L, Valagussa P, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

