Comprehensive meta-QTL analysis for dissecting the genetic architecture of stripe rust resistance in bread wheat
- PMID: 37173660
- PMCID: PMC10182688
- DOI: 10.1186/s12864-023-09336-y
Comprehensive meta-QTL analysis for dissecting the genetic architecture of stripe rust resistance in bread wheat
Abstract
Background: Yellow or stripe rust, caused by the fungus Puccinia striiformis f. sp. tritici (Pst) is an important disease of wheat that threatens wheat production. Since developing resistant cultivars offers a viable solution for disease management, it is essential to understand the genetic basis of stripe rust resistance. In recent years, meta-QTL analysis of identified QTLs has gained popularity as a way to dissect the genetic architecture underpinning quantitative traits, including disease resistance.
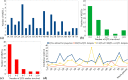
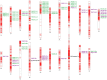
Results: Systematic meta-QTL analysis involving 505 QTLs from 101 linkage-based interval mapping studies was conducted for stripe rust resistance in wheat. For this purpose, publicly available high-quality genetic maps were used to create a consensus linkage map involving 138,574 markers. This map was used to project the QTLs and conduct meta-QTL analysis. A total of 67 important meta-QTLs (MQTLs) were identified which were refined to 29 high-confidence MQTLs. The confidence interval (CI) of MQTLs ranged from 0 to 11.68 cM with a mean of 1.97 cM. The mean physical CI of MQTLs was 24.01 Mb, ranging from 0.0749 to 216.23 Mb per MQTL. As many as 44 MQTLs colocalized with marker-trait associations or SNP peaks associated with stripe rust resistance in wheat. Some MQTLs also included the following major genes- Yr5, Yr7, Yr16, Yr26, Yr30, Yr43, Yr44, Yr64, YrCH52, and YrH52. Candidate gene mining in high-confidence MQTLs identified 1,562 gene models. Examining these gene models for differential expressions yielded 123 differentially expressed genes, including the 59 most promising CGs. We also studied how these genes were expressed in wheat tissues at different phases of development.
Conclusion: The most promising MQTLs identified in this study may facilitate marker-assisted breeding for stripe rust resistance in wheat. Information on markers flanking the MQTLs can be utilized in genomic selection models to increase the prediction accuracy for stripe rust resistance. The candidate genes identified can also be utilized for enhancing the wheat resistance against stripe rust after in vivo confirmation/validation using one or more of the following methods: gene cloning, reverse genetic methods, and omics approaches.
Keywords: Candidate genes; Consensus map; MQTL; Meta-analysis; Stripe rust.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Genome-wide association mapping for stripe rust (Puccinia striiformis F. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.).Theor Appl Genet. 2015 Jun;128(6):1083-101. doi: 10.1007/s00122-015-2492-2. Epub 2015 Mar 10. Theor Appl Genet. 2015. PMID: 25754424
-
Genome-wide DArT and SNP scan for QTL associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in elite ICARDA wheat (Triticum aestivum L.) germplasm.Theor Appl Genet. 2015 Jul;128(7):1277-95. doi: 10.1007/s00122-015-2504-2. Epub 2015 Apr 8. Theor Appl Genet. 2015. PMID: 25851000
-
Meta-QTL analysis and identification of candidate genes for multiple-traits associated with spot blotch resistance in bread wheat.Sci Rep. 2024 Jun 7;14(1):13083. doi: 10.1038/s41598-024-63924-w. Sci Rep. 2024. PMID: 38844568 Free PMC article.
-
Nucleic acid-based strategies to mitigate stripe rust disease of wheat for achieving global food security - A review.Int J Biol Macromol. 2025 Aug;319(Pt 4):145353. doi: 10.1016/j.ijbiomac.2025.145353. Epub 2025 Jun 17. Int J Biol Macromol. 2025. PMID: 40553877 Review.
-
Genome-wide atlas of rust resistance loci in wheat.Theor Appl Genet. 2024 Jul 9;137(8):179. doi: 10.1007/s00122-024-04689-8. Theor Appl Genet. 2024. PMID: 38980436 Free PMC article. Review.
Cited by
-
Unveiling yellow rust resistance in the near-Himalayan region: insights from a nested association mapping study.Theor Appl Genet. 2025 Jun 5;138(7):135. doi: 10.1007/s00122-025-04886-z. Theor Appl Genet. 2025. PMID: 40471307 Free PMC article.
-
Mapping rust resistance in European winter wheat: many QTLs for yellow rust resistance, but only a few well characterized genes for stem rust resistance.Theor Appl Genet. 2024 Sep 5;137(9):215. doi: 10.1007/s00122-024-04731-9. Theor Appl Genet. 2024. PMID: 39235622 Free PMC article.
-
Genome-wide association mapping for the identification of stripe rust resistance loci in US hard winter wheat.Theor Appl Genet. 2025 Mar 10;138(4):67. doi: 10.1007/s00122-025-04858-3. Theor Appl Genet. 2025. PMID: 40063245 Free PMC article.
-
Rapid reprogramming and stabilization of homoeolog expression bias in hexaploid wheat biparental populations.Genome Biol. 2025 May 28;26(1):147. doi: 10.1186/s13059-025-03598-3. Genome Biol. 2025. PMID: 40437599 Free PMC article.
-
Genetic dissection of crown rust resistance in oat and the identification of key adult plant resistance genes.Plant Genome. 2025 Jun;18(2):e70059. doi: 10.1002/tpg2.70059. Plant Genome. 2025. PMID: 40511550 Free PMC article.
References
-
- Afzal SN, Haque MI, Ahmedani MS, Bashir S, Rattu AR. Assessment of yield losses caused by Puccinia striiformis triggering stripe rust in the most common wheat varieties. Pak J Bot. 2017;39:2127–2134.
-
- Pradhan AK, Kumar S, Singh AK, Budhlakoti N, Mishra DC, Chauhan D, Mittal S, Grover M, Kumar S, Gangwar OP, Kumar S. Identification of QTLs/defense genes effective at seedling stage against prevailing races of wheat stripe rust in India. Front Genet. 2020;11:572975. doi: 10.3389/fgene.2020.572975. - DOI - PMC - PubMed
-
- Chen XM. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can J Plant Pathol. 2005;27:314–337. doi: 10.1080/07060660509507230. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

