Bos d 13, A Novel Heat-Stable Beef Allergen
- PMID: 37173826
- PMCID: PMC10909433
- DOI: 10.1002/mnfr.202200601
Bos d 13, A Novel Heat-Stable Beef Allergen
Abstract
Scope: Red meat, a staple food of Western diets, can also induce IgE-mediated allergic reactions. Yet, apart from the heat-labile protein serum albumin and the carbohydrate α-Gal, the molecules causing allergic reactions to red meat remain unknown.
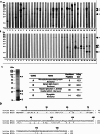
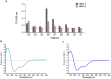
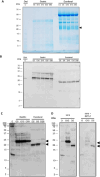
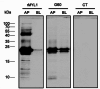
Methods and results: IgE reactivity profiles of beef-sensitized individuals are analyzed by IgE-immunoblotting with protein extracts from raw and cooked beef. Two IgE-reactive proteins are identified by peptide mass fingerprinting as myosinlight chain 1 (MYL1) and myosin light chain 3 (MYL3) in cooked beef extract and are designated Bos d 13 isoallergens. MYL1 and MYL3 are produced recombinantly in Escherichia coli. ELISAs proved their IgE reactivity and circular dichroism analysis showed that they represent folded molecules with remarkable thermal stability. In vitro gastrointestinal digestion experiments showed the higher stability of rMYL1 as compared to rMYL3. Exposure of a monolayer of Caco-2 cells to rMYL1 indicated that the molecule is able to cross intestinal epithelial cells without disturbing the integrity of the tight junctions, suggesting the sensitizing capacity of MYL1.
Conclusion: MYLs are identified as novel heat-stable bovine meat allergens.
Keywords: allergens; heat stability; meat allergy; myosin light chain; recombinant allergens; red meat; stability to digestion.
© 2023 The Authors. Molecular Nutrition & Food Research published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

