PRAME Is a Novel Target of Tumor-Intrinsic Gas6/Axl Activation and Promotes Cancer Cell Invasion in Hepatocellular Carcinoma
- PMID: 37173882
- PMCID: PMC10177160
- DOI: 10.3390/cancers15092415
PRAME Is a Novel Target of Tumor-Intrinsic Gas6/Axl Activation and Promotes Cancer Cell Invasion in Hepatocellular Carcinoma
Abstract
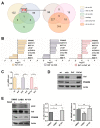
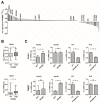
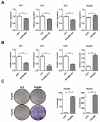
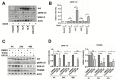
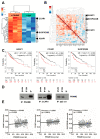
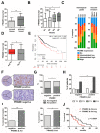
(1) Background: Activation of the receptor tyrosine kinase Axl by Gas6 fosters oncogenic effects in hepatocellular carcinoma (HCC), associating with increased mortality of patients. The impact of Gas6/Axl signaling on the induction of individual target genes in HCC and its consequences is an open issue. (2) Methods: RNA-seq analysis of Gas6-stimulated Axl-proficient or Axl-deficient HCC cells was used to identify Gas6/Axl targets. Gain- and loss-of-function studies as well as proteomics were employed to characterize the role of PRAME (preferentially expressed antigen in melanoma). Expression of Axl/PRAME was assessed in publicly available HCC patient datasets and in 133 HCC cases. (3) Results: Exploitation of well-characterized HCC models expressing Axl or devoid of Axl allowed the identification of target genes including PRAME. Intervention with Axl signaling or MAPK/ERK1/2 resulted in reduced PRAME expression. PRAME levels were associated with a mesenchymal-like phenotype augmenting 2D cell migration and 3D cell invasion. Interactions with pro-oncogenic proteins such as CCAR1 suggested further tumor-promoting functions of PRAME in HCC. Moreover, PRAME showed elevated expression in Axl-stratified HCC patients, which correlates with vascular invasion and lowered patient survival. (4) Conclusions: PRAME is a bona fide target of Gas6/Axl/ERK signaling linked to EMT and cancer cell invasion in HCC.
Keywords: Axl; EMT; Gas6; HCC; PRAME; dedifferentiation; immunotherapy; invasion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Tumor-Extrinsic Axl Expression Shapes an Inflammatory Microenvironment Independent of Tumor Cell Promoting Axl Signaling in Hepatocellular Carcinoma.Int J Mol Sci. 2024 Apr 10;25(8):4202. doi: 10.3390/ijms25084202. Int J Mol Sci. 2024. PMID: 38673795 Free PMC article.
-
Gas6/Axl pathway promotes tumor invasion through the transcriptional activation of Slug in hepatocellular carcinoma.Carcinogenesis. 2014 Apr;35(4):769-75. doi: 10.1093/carcin/bgt372. Epub 2013 Nov 14. Carcinogenesis. 2014. PMID: 24233839
-
Synergism of the receptor tyrosine kinase Axl with ErbB receptors mediates resistance to regorafenib in hepatocellular carcinoma.Front Oncol. 2023 Sep 8;13:1238883. doi: 10.3389/fonc.2023.1238883. eCollection 2023. Front Oncol. 2023. PMID: 37746265 Free PMC article.
-
Gas6/Axl Signaling Pathway in the Tumor Immune Microenvironment.Cancers (Basel). 2020 Jul 9;12(7):1850. doi: 10.3390/cancers12071850. Cancers (Basel). 2020. PMID: 32660000 Free PMC article. Review.
-
Dynamics of Axl Receptor Shedding in Hepatocellular Carcinoma and Its Implication for Theranostics.Int J Mol Sci. 2018 Dec 18;19(12):4111. doi: 10.3390/ijms19124111. Int J Mol Sci. 2018. PMID: 30567378 Free PMC article. Review.
Cited by
-
Survival Benefits of Transarterial Chemoembolization Plus Ablation Therapy in Patients With Intermediate or Advanced Hepatocellular Carcinoma: A Propensity Score Matching Study.Cancer Manag Res. 2025 Mar 4;17:483-497. doi: 10.2147/CMAR.S511364. eCollection 2025. Cancer Manag Res. 2025. PMID: 40060703 Free PMC article.
-
Tumor-Extrinsic Axl Expression Shapes an Inflammatory Microenvironment Independent of Tumor Cell Promoting Axl Signaling in Hepatocellular Carcinoma.Int J Mol Sci. 2024 Apr 10;25(8):4202. doi: 10.3390/ijms25084202. Int J Mol Sci. 2024. PMID: 38673795 Free PMC article.
-
Transcriptome profiles of blastocysts originating from oocytes matured in follicular fluid from preovulatory follicles of greater or lesser maturity.BMC Genomics. 2025 Apr 4;26(1):339. doi: 10.1186/s12864-025-11521-0. BMC Genomics. 2025. PMID: 40186098 Free PMC article.
-
PRAME Updated: Diagnostic, Prognostic, and Therapeutic Role in Skin Cancer.Int J Mol Sci. 2024 Jan 27;25(3):1582. doi: 10.3390/ijms25031582. Int J Mol Sci. 2024. PMID: 38338862 Free PMC article. Review.
-
Tumor Microenvironment as a Therapeutic Target in Melanoma Treatment.Cancers (Basel). 2023 Jun 11;15(12):3147. doi: 10.3390/cancers15123147. Cancers (Basel). 2023. PMID: 37370757 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

