Molecular Basis for the Selectivity of DHA and EPA in Sudlow's Drug Binding Sites in Human Serum Albumin with the Combined Use of NMR and Docking Calculations
- PMID: 37175134
- PMCID: PMC10180286
- DOI: 10.3390/molecules28093724
Molecular Basis for the Selectivity of DHA and EPA in Sudlow's Drug Binding Sites in Human Serum Albumin with the Combined Use of NMR and Docking Calculations
Abstract
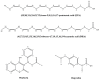
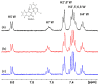
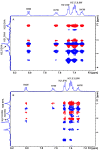
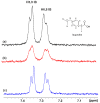
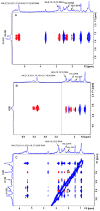
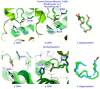
Medium- and long-chain saturated and unsaturated free fatty acids (FFAs) are known to bind to human serum albumin (HSA), the main plasma carrier protein. Atomic-level structural data regarding the binding mode in Sudlow's sites I (FA7) and II (FA4, FA3) of the polyunsaturated ω-3 fatty acids docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), however, are largely unknown. Herein, we report the combined use of saturation transfer difference (STD) and Interligand NOEs for Pharmacophore Mapping (INPHARMA) NMR techniques and molecular docking calculations to investigate the binding mode of DHA and EPA in Sudlow's sites Ι and ΙΙ of HSA. The docking calculations and the significant number of interligand NOEs between DHA and EPA and the drugs warfarin and ibuprofen, which are stereotypical ligands for Sudlow's sites I and II, respectively, were interpreted in terms of competitive binding modes and the presence of two orientations of DHA and EPA at the binding sites FA7 and FA4. The exceptional flexibility of the long-chain DHA and EPA and the formation of strongly folded structural motives are the key properties of HSA-PUFA complexes.
Keywords: DHA; EPA; HSA; INPHARMA NMR; STD NMR; docking calculations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Vance D.E., Vance J.E. Biochemistry of Lipids, Lipoproteins, and Membranes. 5th ed. Elsevier; Amsterdam, The Netherlands: 2008.
-
- Akoh C.C., Min D.B. Food Lipids, Chemistry, Nutrition and Biochemistry. 1st ed. Marcel Dekker Inc.; New York, NY, USA: 2002.
-
- Leray C. Dietary Lipids for Healthy Brain Function. CRC Press; Boca Ratan, FL, USA: 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

