Rapid and Sensitive Detection of Streptococcus iniae in Trachinotus ovatus Based on Multienzyme Isothermal Rapid Amplification
- PMID: 37175440
- PMCID: PMC10178759
- DOI: 10.3390/ijms24097733
Rapid and Sensitive Detection of Streptococcus iniae in Trachinotus ovatus Based on Multienzyme Isothermal Rapid Amplification
Abstract
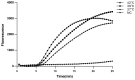
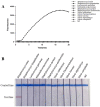
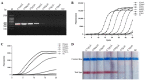
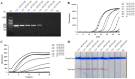
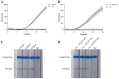
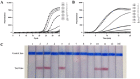
Infectious diseases caused by Streptococcus iniae lead to massive death of fish, compose a serious threat to the global aquaculture industry, and constitute a risk to humans who deal with raw fish. In order to realize the early diagnosis of S. iniae, and control the outbreak and spread of disease, it is of great significance to establish fast, sensitive, and convenient detection methods for S. iniae. In the present study, two methods of real-time MIRA (multienzyme isothermal rapid amplification, MIRA) and MIRA-LFD (combining MIRA with lateral flow dipsticks (LFD)) for the simA gene of S. iniae were established, which could complete amplification at a constant temperature of 42 °C within 20 min. Real-time MIRA and MIRA-LFD assays showed high sensitivity (97 fg/μL or 7.6 × 102 CFU/mL), which were consistent with the sensitivity of real-time PCR and 10 times higher than that of PCR with strong specificity, repeatability simplicity, and rapidity for S. iniae originating from Trachinotus ovatus. In summary, real-time MIRA and MIRA-LFD provide effective ways for early diagnosis of S. iniae in aquaculture, especially for units in poor conditions.
Keywords: Streptococcus iniae; lateral flow dipsticks; multienzyme isothermal rapid amplification; rapid detection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
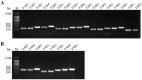
Similar articles
-
Development and evaluation of a rapid and sensitive multienzyme isothermal rapid amplification with a lateral flow dipstick assay for detection of Acinetobacter baumannii in spiked blood specimens.Front Cell Infect Microbiol. 2022 Oct 19;12:1010201. doi: 10.3389/fcimb.2022.1010201. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36339332 Free PMC article.
-
Visible and rapid detection of feline chaphamaparvovirus using multienzyme isothermal rapid amplification and lateral flow dipstick assay.Front Cell Infect Microbiol. 2025 Jan 22;15:1490948. doi: 10.3389/fcimb.2025.1490948. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 39917634 Free PMC article.
-
Establishment of a Rapid Detection Method for Highly Virulent Klebsiella Pneumoniae Based on Multienzyme Isothermal Rapid Amplification Technology.Clin Lab. 2024 Sep 1;70(9). doi: 10.7754/Clin.Lab.2024.240137. Clin Lab. 2024. PMID: 39257119
-
Rapid and sensitive detection of shrimp yellow head virus by loop-mediated isothermal amplification combined with a lateral flow dipstick.J Virol Methods. 2013 Mar;188(1-2):51-6. doi: 10.1016/j.jviromet.2012.11.041. Epub 2012 Dec 7. J Virol Methods. 2013. PMID: 23219929 Review.
-
Loop-mediated isothermal amplification: an emerging technology for detection of fish and shellfish pathogens.J Fish Dis. 2005 Oct;28(10):573-81. doi: 10.1111/j.1365-2761.2005.00670.x. J Fish Dis. 2005. PMID: 16302951 Free PMC article. Review.
Cited by
-
Plasmonic nanoparticle sensors: current progress, challenges, and future prospects.Nanoscale Horiz. 2024 Nov 19;9(12):2085-2166. doi: 10.1039/d4nh00226a. Nanoscale Horiz. 2024. PMID: 39240539 Free PMC article. Review.
-
Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests.Biosensors (Basel). 2023 Sep 15;13(9):885. doi: 10.3390/bios13090885. Biosensors (Basel). 2023. PMID: 37754119 Free PMC article. Review.
-
Development of Multienzyme Isothermal Rapid Amplification (MIRA) Combined with Lateral-Flow Dipstick (LFD) Assay to Detect Species-Specific tlh and Pathogenic trh and tdh Genes of Vibrio parahaemolyticus.Pathogens. 2024 Jan 6;13(1):57. doi: 10.3390/pathogens13010057. Pathogens. 2024. PMID: 38251364 Free PMC article.
-
Development of a multienzyme isothermal and lateral flow dipstick combination assay for the rapid detection of goose astrovirus II.Front Cell Infect Microbiol. 2024 Aug 6;14:1424212. doi: 10.3389/fcimb.2024.1424212. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39165916 Free PMC article.
-
Evaluation and Application of the MIRA-qPCR Method for Rapid Detection of Norovirus Genogroup II in Shellfish.Microorganisms. 2025 Mar 21;13(4):712. doi: 10.3390/microorganisms13040712. Microorganisms. 2025. PMID: 40284551 Free PMC article.
References
-
- Hayat M., Yusoff M., Samad M., Razak I.A., Yasin I., Thompson K., Hasni K. Efficacy of Feed-Based Formalin-Killed Vaccine of Streptococcus iniae Stimulates the Gut-Associated Lymphoid Tissues and Immune Response of Red Hybrid Tilapia. Vaccines. 2021;9:51. doi: 10.3390/vaccines9010051. - DOI - PMC - PubMed
-
- Guo S., Mo Z., Wang Z., Xu J., Li Y., Dan X., Li A. Isolation and pathogenicity of Streptococcus iniae in offshore cage-cultured Trachinotus ovatus in China. Aquaculture. 2018;492:247–252. doi: 10.1016/j.aquaculture.2018.04.015. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

