Gut Microbiome and Small RNA Integrative-Omic Perspective of Meconium and Milk-FED Infant Stool Samples
- PMID: 37175775
- PMCID: PMC10179101
- DOI: 10.3390/ijms24098069
Gut Microbiome and Small RNA Integrative-Omic Perspective of Meconium and Milk-FED Infant Stool Samples
Abstract
The human gut microbiome plays an important role in health, and its initial development is conditioned by many factors, such as feeding. It has also been claimed that this colonization is guided by bacterial populations, the dynamic virome, and transkingdom interactions between host and microbial cells, partially mediated by epigenetic signaling. In this article, we characterized the bacteriome, virome, and smallRNome and their interaction in the meconium and stool samples from infants. Bacterial and viral DNA and RNA were extracted from the meconium and stool samples of 2- to 4-month-old milk-fed infants. The bacteriome, DNA and RNA virome, and smallRNome were assessed using 16S rRNA V4 sequencing, viral enrichment sequencing, and small RNA sequencing protocols, respectively. Data pathway analysis and integration were performed using the R package mixOmics. Our findings showed that the bacteriome differed among the three groups, while the virome and smallRNome presented significant differences, mainly between the meconium and stool of milk-fed infants. The gut environment is rapidly acquired after birth, and it is highly adaptable due to the interaction of environmental factors. Additionally, transkingdom interactions between viruses and bacteria can influence host and smallRNome profiles. However, virome characterization has several protocol limitations that must be considered.
Keywords: bacteriome; breast-fed; formula-fed; gut microbiota; holobiont; meconium; metagenomics; multiomics; smallRNome; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


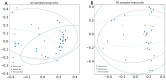
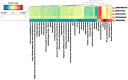


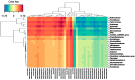
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

