CLEC16A-An Emerging Master Regulator of Autoimmunity and Neurodegeneration
- PMID: 37175930
- PMCID: PMC10179542
- DOI: 10.3390/ijms24098224
CLEC16A-An Emerging Master Regulator of Autoimmunity and Neurodegeneration
Abstract
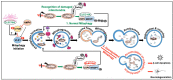
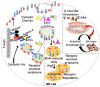
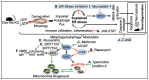
CLEC16A is emerging as an important genetic risk factor for several autoimmune disorders and for Parkinson disease (PD), opening new avenues for translational research and therapeutic development. While the exact role of CLEC16A in health and disease is still being elucidated, the gene plays a critical role in the regulation of autophagy, mitophagy, endocytosis, intracellular trafficking, immune function, and in biological processes such as insulin secretion and others that are important to cellular homeostasis. As shown in both human and animal modeling studies, CLEC16A hypofunction predisposes to both autoinflammatory phenotype and neurodegeneration. While the two are clearly related, further functional studies are needed to fully understand the mechanisms involved for optimized therapeutic interventions. Based on recent data, mitophagy-inducing drugs may be warranted, and such therapy should be tested in clinical trials as these drugs would tackle the underlying pathogenic mechanism (s) and could treat or prevent symptoms of autoimmunity and neurodegeneration in individuals with CLEC16A risk variants. Accordingly, interventions directed at reversing the dysregulated mitophagy and the consequences of loss of function of CLEC16A without activating other detrimental cellular pathways could present an effective therapy. This review presents the emerging role of CLEC16A in health and disease and provides an update on the disease processes that are attributed to variants located in the CLEC16A gene, which are responsible for autoimmune disorders and neurodegeneration with emphasis on how this information is being translated into practical and effective applications in the clinic.
Keywords: C-type lectin-like domain family 16A (CLEC16A) gene; CLEC16A; Parkinson’s disease (PD); autoimmunity; autophagy; genome-wide association studies (GWAS); mitophagy; neurodegeneration; suppressor of cytokine signaling 1 (SOCS1); susceptibility loci; type 1 diabetes (T1D).
Conflict of interest statement
Hakon Hakonarson, Marina Bakay and Rahul Pandey report they have three patents pending on ‘New innovative weight reduction therapies targeting CLEC16A’, ‘Role of CLEC16A and SOCS1 as therapeutic options in autoimmunity modeled through a UBC-Cre-CLEC16AloxP Phenotype Mouse’ and ‘Probucol ameliorates the autoimmune, lipodystrophic and neurodegenerative phenotypes in CLEC16A KO mice’.
Figures


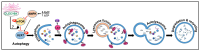
References
-
- Schuster C., Gerold K.D., Schober K., Probst L., Boerner K., Kim M.J., Ruckdeschel A., Serwold T., Kissler S. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity. 2015;42:942–952. doi: 10.1016/j.immuni.2015.04.011. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

