Clostridia and Enteroviruses as Synergistic Triggers of Type 1 Diabetes Mellitus
- PMID: 37176044
- PMCID: PMC10179352
- DOI: 10.3390/ijms24098336
Clostridia and Enteroviruses as Synergistic Triggers of Type 1 Diabetes Mellitus
Abstract
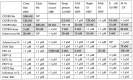
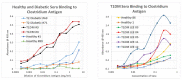
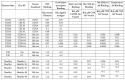
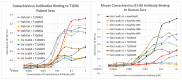
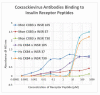
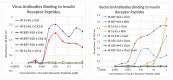
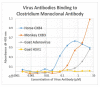
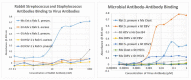
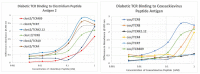
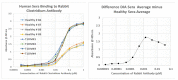
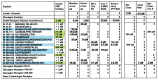
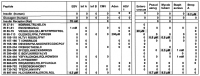
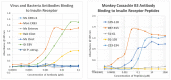
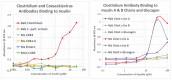
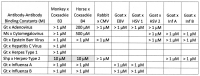
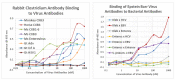
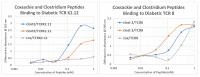
What triggers type 1 diabetes mellitus (T1DM)? One common assumption is that triggers are individual microbes that mimic autoantibody targets such as insulin (INS). However, most microbes highly associated with T1DM pathogenesis, such as coxsackieviruses (COX), lack INS mimicry and have failed to induce T1DM in animal models. Using proteomic similarity search techniques, we found that COX actually mimicked the INS receptor (INSR). Clostridia were the best mimics of INS. Clostridia antibodies cross-reacted with INS in ELISA experiments, confirming mimicry. COX antibodies cross-reacted with INSR. Clostridia antibodies further bound to COX antibodies as idiotype-anti-idiotype pairs conserving INS-INSR complementarity. Ultraviolet spectrometry studies demonstrated that INS-like Clostridia peptides bound to INSR-like COX peptides. These complementary peptides were also recognized as antigens by T cell receptor sequences derived from T1DM patients. Finally, most sera from T1DM patients bound strongly to inactivated Clostridium sporogenes, while most sera from healthy individuals did not; T1DM sera also exhibited evidence of anti-idiotype antibodies against idiotypic INS, glutamic acid decarboxylase, and protein tyrosine phosphatase non-receptor (islet antigen-2) antibodies. These results suggest that T1DM is triggered by combined enterovirus-Clostridium (and possibly combined Epstein-Barr-virus-Streptococcal) infections, and the probable rate of such co-infections approximates the rate of new T1DM diagnoses.
Keywords: COX; Clostridium; T cell receptors; circulating immune complexes; complementary antigens; diabetes; idiotype–anti-idiotype; synergism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













Similar articles
-
T-Cell Receptor Sequences Identify Combined Coxsackievirus-Streptococci Infections as Triggers for Autoimmune Myocarditis and Coxsackievirus-Clostridia Infections for Type 1 Diabetes.Int J Mol Sci. 2024 Feb 1;25(3):1797. doi: 10.3390/ijms25031797. Int J Mol Sci. 2024. PMID: 38339075 Free PMC article.
-
Enterovirus infection may induce humoral immune response reacting with islet cell autoantigens in humans.J Med Virol. 2003 Mar;69(3):426-40. doi: 10.1002/jmv.10306. J Med Virol. 2003. PMID: 12526055
-
Cross-reactive peptide epitopes of Enterovirus Coxsackie B4 and human glutamic acid decarboxylase detecting antibodies in latent autoimmune diabetes in adults versus type 1 diabetes.Clin Chim Acta. 2021 Apr;515:73-79. doi: 10.1016/j.cca.2021.01.002. Epub 2021 Jan 7. Clin Chim Acta. 2021. PMID: 33422493
-
Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host.Nat Rev Endocrinol. 2010 May;6(5):279-89. doi: 10.1038/nrendo.2010.27. Epub 2010 Mar 30. Nat Rev Endocrinol. 2010. PMID: 20351698 Review.
-
Do microbes have a causal role in type 1 diabetes?Med Sci Monit. 2005 Mar;11(3):RA63-9. Med Sci Monit. 2005. PMID: 15735577 Review.
Cited by
-
T-Cell Receptor Sequences Identify Combined Coxsackievirus-Streptococci Infections as Triggers for Autoimmune Myocarditis and Coxsackievirus-Clostridia Infections for Type 1 Diabetes.Int J Mol Sci. 2024 Feb 1;25(3):1797. doi: 10.3390/ijms25031797. Int J Mol Sci. 2024. PMID: 38339075 Free PMC article.
References
-
- Krischer J.P., Liu X., Lernmark Å., Hagopian W.A., Rewers M.J., She J.X., Toppari J., Ziegler A.G., Akolkar B., TEDDY Study Group Predictors of the Initiation of Islet Autoimmunity and Progression to Multiple Autoantibodies and Clinical Diabetes: The TEDDY Study. Diabetes Care. 2022;45:2271–2281. doi: 10.2337/dc21-2612. - DOI - PMC - PubMed
-
- Bauer W., Veijola R., Lempainen J., Kiviniemi M., Härkönen T., Toppari J., Knip M., Gyenesei A., Ilonen J. Age at Seroconversion, HLA Genotype, and Specificity of Autoantibodies in Progression of Islet Autoimmunity in Childhood. J. Clin. Endocrinol. Metab. 2019;104:4521–4530. doi: 10.1210/jc.2019-00421. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

