Transcriptomic Analysis on the Peel of UV-B-Exposed Peach Fruit Reveals an Upregulation of Phenolic- and UVR8-Related Pathways
- PMID: 37176875
- PMCID: PMC10180693
- DOI: 10.3390/plants12091818
Transcriptomic Analysis on the Peel of UV-B-Exposed Peach Fruit Reveals an Upregulation of Phenolic- and UVR8-Related Pathways
Abstract
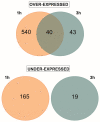
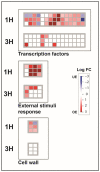
UV-B treatment deeply influences plant physiology and biochemistry, especially by activating the expression of responsive genes involved in UV-B acclimation through a UV-B-specific perception mechanism. Although the UV-B-related molecular responses have been widely studied in Arabidopsis, relatively few research reports deepen the knowledge on the influence of post-harvest UV-B treatment on fruit. In this work, a transcriptomic approach is adopted to investigate the transcriptional modifications occurring in the peel of UV-B-treated peach (Prunus persica L., cv Fairtime) fruit after harvest. Our analysis reveals a higher gene regulation after 1 h from the irradiation (88% of the differentially expressed genes-DEGs), compared to 3 h recovery. The overexpression of genes encoding phenylalanine ammonia-lyase (PAL), chalcone syntase (CHS), chalcone isomerase (CHI), and flavonol synthase (FLS) revealed a strong activation of the phenylpropanoid pathway, resulting in the later increase in the concentration of specific flavonoid classes, e.g., anthocyanins, flavones, dihydroflavonols, and flavanones, 36 h after the treatment. Upregulation of UVR8-related genes (HY5, COP1, and RUP) suggests that UV-B-triggered activation of the UVR8 pathway occurs also in post-harvest peach fruit. In addition, a regulation of genes involved in the cell-wall dismantling process (PME) is observed. In conclusion, post-harvest UV-B exposure deeply affects the transcriptome of the peach peel, promoting the activation of genes implicated in the biosynthesis of phenolics, likely via UVR8. Thus, our results might pave the way to a possible use of post-harvest UV-B treatments to enhance the content of health-promoting compounds in peach fruits and extending the knowledge of the UVR8 gene network.
Keywords: Prunus persica; UV-B radiation; flavonoids; metabolomics; secondary metabolism; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Beyond the Visible and Below the Peel: How UV-B Radiation Influences the Phenolic Profile in the Pulp of Peach Fruit. A Biochemical and Molecular Study.Front Plant Sci. 2020 Oct 30;11:579063. doi: 10.3389/fpls.2020.579063. eCollection 2020. Front Plant Sci. 2020. PMID: 33193522 Free PMC article.
-
Comparative "phenol-omics" and gene expression analyses in peach (Prunus persica) skin in response to different postharvest UV-B treatments.Plant Physiol Biochem. 2019 Feb;135:511-519. doi: 10.1016/j.plaphy.2018.11.009. Epub 2018 Nov 12. Plant Physiol Biochem. 2019. PMID: 30463801
-
UV-B exposure reduces the activity of several cell wall-dismantling enzymes and affects the expression of their biosynthetic genes in peach fruit (Prunus persica L., cv. Fairtime, melting phenotype).Photochem Photobiol Sci. 2019 May 15;18(5):1280-1289. doi: 10.1039/c8pp00505b. Photochem Photobiol Sci. 2019. PMID: 30907896
-
Perception and Signaling of Ultraviolet-B Radiation in Plants.Annu Rev Plant Biol. 2021 Jun 17;72:793-822. doi: 10.1146/annurev-arplant-050718-095946. Epub 2021 Feb 26. Annu Rev Plant Biol. 2021. PMID: 33636992 Review.
-
Beyond Arabidopsis: Differential UV-B Response Mediated by UVR8 in Diverse Species.Front Plant Sci. 2019 Jun 18;10:780. doi: 10.3389/fpls.2019.00780. eCollection 2019. Front Plant Sci. 2019. PMID: 31275337 Free PMC article. Review.
Cited by
-
WRKY Transcription Factors Modulate the Flavonoid Pathway of Rhododendron chrysanthum Pall. Under UV-B Stress.Plants (Basel). 2025 Jan 4;14(1):133. doi: 10.3390/plants14010133. Plants (Basel). 2025. PMID: 39795393 Free PMC article.
-
Characterization of ZAT12 protein from Prunus persica: role in fruit chilling injury tolerance and identification of gene targets.Planta. 2024 Dec 13;261(1):14. doi: 10.1007/s00425-024-04593-x. Planta. 2024. PMID: 39672956
-
Rhododendron chrysanthum's Primary Metabolites Are Converted to Phenolics More Quickly When Exposed to UV-B Radiation.Biomolecules. 2023 Nov 24;13(12):1700. doi: 10.3390/biom13121700. Biomolecules. 2023. PMID: 38136571 Free PMC article.
-
Exploring large-scale gene coexpression networks in peach (Prunus persica L.): a new tool for predicting gene function.Hortic Res. 2024 Jan 2;11(2):uhad294. doi: 10.1093/hr/uhad294. eCollection 2024 Feb. Hortic Res. 2024. PMID: 38487296 Free PMC article.
-
The Multifaceted Responses of Plants to Visible and Ultraviolet Radiation.Plants (Basel). 2024 Feb 20;13(5):572. doi: 10.3390/plants13050572. Plants (Basel). 2024. PMID: 38475419 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

