Pea Aphid (Acyrthosiphon pisum) Host Races Reduce Heat-Induced Forisome Dispersion in Vicia faba and Trifolium pratense
- PMID: 37176952
- PMCID: PMC10181200
- DOI: 10.3390/plants12091888
Pea Aphid (Acyrthosiphon pisum) Host Races Reduce Heat-Induced Forisome Dispersion in Vicia faba and Trifolium pratense
Abstract
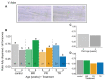
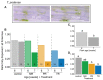
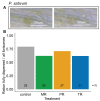
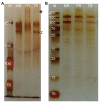
Although phloem-feeding insects such as aphids can cause significant damage to plants, relatively little is known about early plant defenses against these insects. As a first line of defense, legumes can stop the phloem mass flow through a conformational change in phloem proteins known as forisomes in response to Ca2+ influx. However, specialized phloem-feeding insects might be able to suppress the conformational change of forisomes and thereby prevent sieve element occlusion. To investigate this possibility, we triggered forisome dispersion through application of a local heat stimulus to the leaf tips of pea (Pisum sativum), clover (Trifolium pratense) and broad bean (Vicia faba) plants infested with different pea aphid (Acyrthosiphon pisum) host races and monitored forisome responses. Pea aphids were able to suppress forisome dispersion, but this depended on the infesting aphid host race, the plant species, and the age of the plant. Differences in the ability of aphids to suppress forisome dispersion may be explained by differences in the composition and quantity of the aphid saliva injected into the plant. Various mechanisms of how pea aphids might suppress forisome dispersion are discussed.
Keywords: Pisum sativum; Trifolium pratense; Vicia faba; aphid saliva; calcium; legume; pea aphid host race; phloem located defense; phloem protein; sieve element occlusion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ponder K.L., Pritchard J., Harrington R., Bale J.S. Difficulties in location and acceptance of phloem sap combined with reduced concentration of phloem amino acids explain lowered performance of the aphid Rhopalosiphum padi on nitrogen deficient barley (Hordeum vulgare) seedlings. Entomol. Exp. Appl. 2000;97:203–210. doi: 10.1046/j.1570-7458.2000.00731.x. - DOI
-
- Comadira G., Rasool B., Karpinska B., Morris J., Verrall S.R., Hedley P.E., Foyer C.H., Hancock R.D. Nitrogen deficiency in barley (Hordeum vulgare) seedlings induces molecular and metabolic adjustments that trigger aphid resistance. J. Exp. Bot. 2015;66:3639–3655. doi: 10.1093/jxb/erv276. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

